[ Program Manual | User's Guide | Data Files | Databases ]
HelicalWheel plots a peptide sequence as a helical wheel to help you recognize amphiphilic regions.
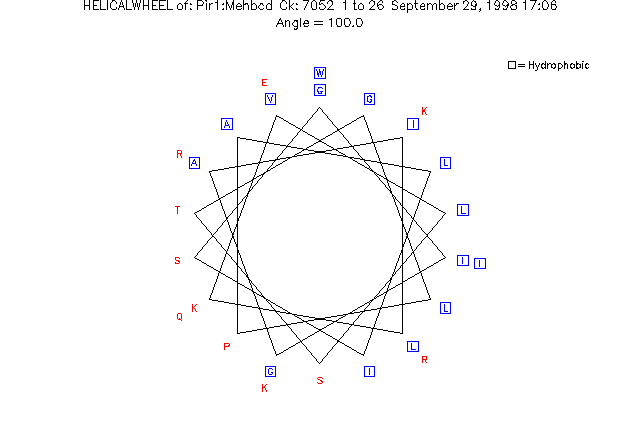
HelicalWheel plots a helical wheel representation of a peptide sequence. Each residue is offset from the preceding one by 100 degrees, the typical angle of rotation for an alpha-helix. The angle of rotation can be changed using -ANGle and -BETa.
Here is a session using HelicalWheel to plot a helical wheel representation of PIR:Mehbcd:
% helicalwheel
HELICALWHEEL of what protein sequence ? PIR:Mehbcd
Begin (* 1 *) ?
End (* 26 *) ?
When your LaserWriter attached to tty07 is ready, press <Return>.
%
If you are reading the Program Manual, the plot from this session is shown in the figure below.

HelicalWheel accepts a single protein sequence as input. If HelicalWheel rejects your protein sequence, see Appendix VI for information on how to change or set the type of a sequence.
Moment makes a contour plot of the helical hydrophobic moment of a peptide sequence. PepPlot shows the maximum hydrophobic moment over the range from 95 to 105 degrees (alpha-helix) and from 159 to 161 degrees (beta-sheet) calculated in a window of six residues.
The Wisconsin Package must be configured for graphics before you run any program with graphics output! If the % setplot command is available in your installation, this is the easiest way to establish your graphics configuration, but you can also use commands like % postscript that correspond to the graphics languages the Wisconsin Package supports. See Chapter 5, Using Graphics in the User's Guide for more information about configuring your process for graphics.
If you need to stop this program, use <Ctrl>C to reset your terminal and session as gracefully as possible. Searches and comparisons write out the results from the part of the search that is complete when you use <Ctrl>C. The graphics device should stop plotting the current page and start plotting the next page. If the current page is the last page, plotters should put the pen away and graphic terminals should return to interactive mode.
All parameters for this program may be added to the command line. Use -CHEck to view the summary below and to specify parameters before the program executes. In the summary below, the capitalized letters in the parameter names are the letters that you must type in order to use the parameter. Square brackets ([ and ]) enclose parameter values that are optional. For more information, see "Using Program Parameters" in Chapter 3, Using Programs in the User's Guide.
Minimal Syntax: % helicalwheel [-INfile=]PIR:Mehbcd -Default Prompted Parameters: -BEGin=1 -END=26 sets the range of interest Local Data Files: -DATa=helicalwheel.dat assigns file of charges and colors for each a.a. -SIMplify=simplify.txt simplifies sequence (See SIMPLIFY) Optional Parameters: -ANGle=100 sets rotation per residue on wheel -BETa sets rotation per residue to 160 degrees -RADius=30 sets radius of wheel to 30 platen units -STARtposition=0 sets angle of first residue on wheel -THRee labels wheel with three letter a.a. codes -NOCHArge suppresses box around hydrophobic residues All GCG graphics programs accept these and other switches. See the Using Graphics chapter of the USERS GUIDE for descriptions. -FIGure[=FileName] stores plot in a file for later input to FIGURE -FONT=3 draws all text on the plot using font 3 -COLor=1 draws entire plot with pen in stall 1 -SCAle=1.2 enlarges the plot by 20 percent (zoom in) -XPAN=10.0 moves plot to the right 10 platen units (pan right) -YPAN=10.0 moves plot up 10 platen units (pan up) -PORtrait rotates plot 90 degrees
The files described below supply auxiliary data to this program. The program automatically reads them from a public data directory unless you either 1) have a data file with exactly the same name in your current working directory; or 2) name a file on the command line with an expression like -DATa1=myfile.dat. For more information see Chapter 4, Using Data Files in the User's Guide.
HelicalWheel reads the file helicalwheel.dat for the hydrophobicity/hydrophilicity and display color of each amino acid.
If you use -SIMplify, HelicalWheel reads the local data file simplify.txt to find the symbol equivalences you want to use. You can specify a simplification table with another name on the command line with an expression like -SIMplify=mysimplify.txt. There is more on the subject of simplification in the documentation for the Simplify program.
You can use the Fetch program to copy and modify these files to suit your own needs.
You can set the parameters listed below from the command line. For more information, see "Using Program Parameters" in Chapter 3, Using Programs in the User's Guide.
simplifies the sequence according to a table of equivalences in the local data file called simplfiy.txt.
sets the angle of rotation per residue on the wheel.
sets the angle of rotation to be 160 degrees (for beta-structure).
sets the radius of the wheel in platen units.
sets the angle of the first residue on the wheel (in degrees clockwise from 12 o'clock).
labels the wheel with three-letter amino acid codes.
suppresses plotting boxes around hydrophobic single-letter residue codes.
The parameters below apply to all Wisconsin Package graphics programs. These and many others are described in detail in Chapter 5, Using Graphics of the User's Guide.
writes the plot as a text file of plotting instructions suitable for input to the Figure program instead of sending it to the device specified in your graphics configuration.
draws all text characters on the plot using Font 3 (see Appendix I).
draws the entire plot with the pen in stall 1.
The parameters below let you expand or reduce the plot (zoom), move it in either direction (pan), or rotate it 90 degrees (rotate).
expands the plot by 20 percent by resetting the scaling factor (normally 1.0) to 1.2 (zoom in). You can expand the axes independently with -XSCAle and -YSCAle. Numbers less than 1.0 contract the plot (zoom out).
moves the plot to the right by 30 platen units (pan right).
moves the plot up by 30 platen units (pan up).
rotates the plot 90 degrees. Usually, plots are displayed with the horizontal axis longer than the vertical (landscape). Note that plots are reduced or enlarged, depending on the platen size, to fill the page.
[ Program Manual | User's Guide | Data Files | Databases ]
Documentation Comments: doc-comments@gcg.com
Technical Support: help@gcg.com
Copyright (c) 1982, 1983, 1985, 1986, 1987, 1989, 1991, 1994, 1995, 1996, 1997, 1998 Genetics Computer Group Inc., a wholly owned subsidiary of Oxford Molecular Group, Inc. All rights reserved.
Licenses and Trademarks Wisconsin Package is a trademark of Genetics Computer Group, Inc. GCG and the GCG logo are registered trademarks of Genetics Computer Group, Inc.
All other product names mentioned in this documentation may be trademarks, and if so, are trademarks or registered trademarks of their respective holders and are used in this documentation for identification purposes only.