
[ Program Manual | User's Guide | Data Files | Databases ]
PlotStructure plots the measures of protein secondary structure in the output file from PeptideStructure. The measures can be shown on parallel panels of a graph or with a two-dimensional "squiggly" representation.
PeptideStructure writes a file with several kinds of protein secondary structure predictions shown as a table. PlotStructure reads the PeptideStructure output file and displays it in any of several different ways.
There are two main options, each with several suboptions:
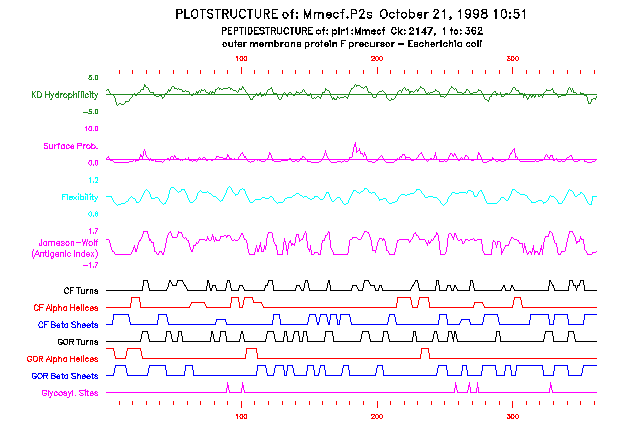
One main option for PlotStructure creates a one-dimensional, multi-paneled plot. The residues are numbered on the x-axis and the attributes are represented as continuous curves in each of several different panels. The horizontal line across the surface probability panel at position 1.0 on the y-axis indicates the expected surface probability calculated for a random sequence. Values above this line indicate an increased probability of being found on the protein surface. The surface probability values are clipped at a height of 10.0 on the y-axis.
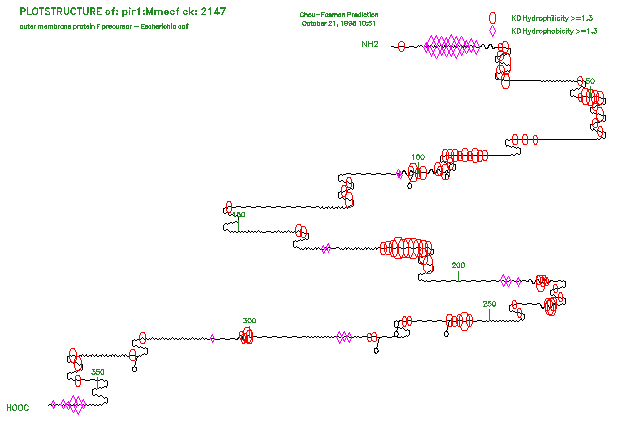
The other main option generates a two-dimensional plot representing predicted secondary structures (alpha-helices, beta-sheets, turns, and coils) with different wave forms. Helices are shown with a sine wave, beta-sheets with a sharp saw-tooth wave, turns with 180 degree turns, and coils with a dull saw-tooth wave. Any of four different quantitative attributes (hydrophilicity, surface probability, flexibility, or antigenic index) can be superimposed over the wave with special symbols wherever the attribute exceeds some set threshold. The size of the symbols is proportional to the value of the attribute. In addition, possible glycosylation sites can be marked on the two-dimensional plot.
Here is a session using PlotStructure to plot the protein secondary structure measures for PIR:Mmecf, which is calculated in the Program Manual example for PeptideStructure:
% plotstructure
PLOTSTRUCTURE of what PEPTIDESTRUCTURE output file ? mmecf.p2s
Plot Begin (* 1 *) ?
Plot End (* 362 *) ?
Do you want a
1)-dimensional (panel graph) or a
2)-dimensional (squiggly) plot
Please choose one (* 1 *):
When your LaserWriter attached to tty07 is ready, press <Return>.
%
If you are reading the Program Manual, the plot from this session and from a session selecting the two-dimentional plot for the same input data are shown in the two figures at the end of this program entry.
PeptideStructure calculates the secondary structure of a peptide and writes a file suitable for input to PlotStructure. PepPlot plots measures of protein secondary structure as a set of parallel curves in different panels of the same plot.
The input file for PlotStructure must be the output file from PeptideStructure. There are some format requirements for this file discussed in the INPUT FILE topic below.
The input file for PlotStructure is the output file from PeptideStructure. PlotStructure reads the first two lines of the input file to identify and document the sequence that was analyzed by PeptideStructure. It also searches for a line with the string Date: for the date of the session with PeptideStructure. This date is shown on all plots so that plots from the same input file may be associated. Here is some of the input file used for the example session:
PEPTIDESTRUCTURE of: pir1:Mmecf check: 2147 from: 1 to: 362
outer membrane protein F precursor - Escherichia coli
Hydrophilicity (Kyte-Doolittle) averaged over a window of: 7
Surface Probability according to Emini
Chain Flexibility according to Karplus-Schulz
Secondary Structure according to Chou-Fasman
Secondary Structure according to Garnier-Osguthorpe-Robson
Antigenicity Index according to Jameson-Wolf
Date: October 21, 1998 10:51
Pos AA GlycoS HyPhil SurfPr FlexPr CF-Pred GORPred AI-Ind ..
1 M . 1.150 1.437 1.000 . H 0.900
2 M . 1.620 1.808 1.000 . H 0.900
3 K . 0.600 0.991 1.000 . H 0.750
///////////////////////////////////////////////////////////////
360 Y . -1.050 0.358 1.000 B B -0.450
361 Q . -1.340 0.653 1.000 B B -0.450
362 F . -0.550 1.125 1.000 B B -0.300
The Wisconsin Package must be configured for graphics before you run any program with graphics output! If the % setplot command is available in your installation, this is the easiest way to establish your graphics configuration, but you can also use commands like % postscript that correspond to the graphics languages the Wisconsin Package supports. See Chapter 5, Using Graphics in the User's Guide for more information about configuring your process for graphics.
If you need to stop this program, use <Ctrl>C to reset your terminal and session as gracefully as possible. Searches and comparisons write out the results from the part of the search that is complete when you use <Ctrl>C. The graphics device should stop plotting the current page and start plotting the next page. If the current page is the last page, plotters should put the pen away and graphic terminals should return to interactive mode.
All parameters for this program may be added to the command line. Use -CHEck to view the summary below and to specify parameters before the program executes. In the summary below, the capitalized letters in the parameter names are the letters that you must type in order to use the parameter. Square brackets ([ and ]) enclose parameter values that are optional. For more information, see "Using Program Parameters" in Chapter 3, Using Programs in the User's Guide.
Minimal Syntax: % plotstructure [-INfile=]mmecf.p2s -Default
Prompted Parameters:
-BEGin=1 -END=362 sets first and last residue to plot
-MENu1=1 plots 1)-dimensional (panel) or 2)-dimensional (squiggly)
-MENu2=c plots either C)hou-Fasman or G)arnier-Robson prediction
-MENu3=h superimposes: H)ydrophilicity, S)urface-probability,
F)lexibility, A)ntigenicity or N)othing
Local Data Files: None
Optional Parameters:
-THReshold=1.3 sets threshold for superimposed measure (Menu3)
-NOSTRONgonly displays both strong and weak Chou-Fasman structures
-NOGLYCosylation suppresses the display of possible glycosylation sites
-NUMbering=50 numbers plot in intervals of 50 aa
-DENsity=1000 sets the number of residues per 100 platen units
(1-dimensional plot, only)
All GCG graphics programs accept these and other switches. See the Using
Graphics chapter of the USERS GUIDE for descriptions.
-FIGure[=FileName] stores plot in a file for later input to FIGURE
-FONT=3 draws all text on the plot using font 3
-COLor=1 draws entire plot with pen in stall 1
-SCAle=1.2 enlarges the plot by 20 percent (zoom in)
-XPAN=10.0 moves plot to the right 10 platen units (pan right)
-YPAN=10.0 moves plot up 10 platen units (pan up)
-PORtrait rotates plot 90 degrees
PeptideStructure and PlotStructure were communicated to us by Dr. Hans Wolf for Drs. Susanne Modrow and Manfred Motz of the Max von Pettenkofer-Institut of the University of Munich and for Bradford Jameson of the California Institute of Technology. The programs were written for them by Dr. B. J. Foertsch and G. Herrmann and were modified for compatibility with GCG Version 5 by John Devereux. The graphic schematic and overlying symbols for hydropathy and glycosylation are based on the early work (1983) of Ellis Golub of the University of Pennsylvania. These programs have been described and used by Cohen et al. (J. Virol. 49; 102-108 (1984)), Starcich et al. (Cell, 45; 637-648 (1986)), Motz et al. (Gene, 42; 303-312 (1986)) and by Modrow and Wolf (Proc. Natl. Acad. Sci. USA 83; 5703-5707 (1986)). The method for calculating antigenic index is described by Jameson and Wolf (CABIOS, 4(1), 181-186 (1988)). Any use of PeptideStructure or PlotStructure for published research should cite the Jameson and Wolf article.
None.
You can set the parameters listed below from the command line. For more information, see "Using Program Parameters" in Chapter 3, Using Programs in the User's Guide.
sets the type of plot: a one-dimensional (panel) plot (1) or a two-dimensional (squiggly) plot (2).
sets the type of secondary structure prediction to use: Chou-Fasman (C) or Garnier-Robson (G).
superimposes one of the following over the "squiggly" plot: hydrophilicity (H), surface-probability (S), flexibility (F), antigenicity (A) or superimposes none of these (N).
sets the threshold for the display of the superimposed statistic (hydrophilicity, surface probability, flexibility, or antigenicity) selected with -MENu3 when you are plotting the "squiggly" plot.
Usually, PlotStructure only shows the Chou-Fasman predictions that are strong. These predictions are shown as capital letters in the PeptideStructure output file. You can plot the weakly predicted features with -NOSTRONgonly.
suppresses the display of possible glycosylation sites.
For the two-dimensional plot, numbering is usually enabled with a numbering interval of 50 residues. You can assign a different value to the parameter or you can suppress numbering altogether with -NONUMbering.
sets the number of bases or amino acids per 100 platen units (PU). This is usually equivalent to the number of bases or amino acids per page. Output from different GCG graphics programs that are run at the same density can be compared by lining up the plots on a light box.
Use -DENsity to change the density of the one-dimensional (panel) plot; PlotStructure ignores this parameter when you create a two-dimensional (squiggly) plot.
The parameters below apply to all Wisconsin Package graphics programs. These and many others are described in detail in Chapter 5, Using Graphics of the User's Guide.
writes the plot as a text file of plotting instructions suitable for input to the Figure program instead of sending it to the device specified in your graphics configuration.
draws all text characters on the plot using Font 3 (see Appendix I).
draws the entire plot with the pen in stall 1.
The parameters below let you expand or reduce the plot (zoom), move it in either direction (pan), or rotate it 90 degrees (rotate).
expands the plot by 20 percent by resetting the scaling factor (normally 1.0) to 1.2 (zoom in). You can expand the axes independently with -XSCAle and -YSCAle. Numbers less than 1.0 contract the plot (zoom out).
moves the plot to the right by 30 platen units (pan right).
moves the plot up by 30 platen units (pan up).
rotates the plot 90 degrees. Usually, plots are displayed with the horizontal axis longer than the vertical (landscape). Note that plots are reduced or enlarged, depending on the platen size, to fill the page.
[ Program Manual | User's Guide | Data Files | Databases ]
Documentation Comments: doc-comments@gcg.com
Technical Support: help@gcg.com
Copyright (c) 1982, 1983, 1985, 1986, 1987, 1989, 1991, 1994, 1995, 1996, 1997, 1998 Genetics Computer Group Inc., a wholly owned subsidiary of Oxford Molecular Group, Inc. All rights reserved.
Licenses and Trademarks Wisconsin Package is a trademark of Genetics Computer Group, Inc. GCG and the GCG logo are registered trademarks of Genetics Computer Group, Inc.
All other product names mentioned in this documentation may be trademarks, and if so, are trademarks or registered trademarks of their respective holders and are used in this documentation for identification purposes only.