
[ Program Manual | User's Guide | Data Files | Databases ]
SeqEd is an interactive editor for entering and modifying sequences and for assembling parts of existing sequences into new genetic constructs. You can enter sequences from the keyboard or from a digitizer.
SeqEd uses the screen of your terminal as a window into a sequence. It works like the UNIX vi text editor. Changes you make in the sequence take place at the cursor position and are reflected immediately on the screen. You can insert or delete symbols, move the cursor, search for patterns, check sequences by reentering them, and edit documentation and embedded comments.
SetKeys lets you change the positions of the keys on your terminal keyboard to make it more convenient to enter the letters G, A, T, and C.
You can enter a sequence and control SeqEd either from the terminal keyboard or from a Graf/Bar digitizer.
If you are already familiar with the UNIX vi editor, you can learn to use SeqEd quickly. When you run SeqEd with a command like % seqed sample.seq, your screen will look something like this:
sample.seq ***** K E Y B O A R D ***** seqed
: Some documentary text about your sequence can be placed :
: here in the heading. :
: You can have as many lines of header comments as you wish, and :
: you can edit them in this space with the HEAding command. :
1 $The scale below has extra dots where comments occur$
9 <The number indicates which symbol the comment precedes.<
20 >There can be as many comments as you like>
28 $The four comments closest to the cursor are displayed$
AGTCTTAGTCGATCGTAcTGCATRCGA
....|:.......:|.........i.......:.|.........|.........|.........|.........|..
0 10 20 30 40 50 60 70
~~~~~~~~~~~~~~~~~~~^
|......|......|......|......|......|......|......|......|......|......|
0 10 20 30 40 50 60 70 80 90 100
"sample.seq" 27 nucleotides
If you name a sequence file that already exists, SeqEd displays the first four lines of documentation on the top of the screen followed by up to four embedded comments and the base number with which each is associated. SeqEd shows the end of the sequence across the middle of the screen.
If the sequence you name does not exist, SeqEd starts in Heading Mode (see below) to allow you to enter documentation for the new sequence. Use <Ctrl>D to stop editing the documentation.
In Screen Mode the cursor shows your position in the sequence. You can move around in the sequence, add symbols, delete symbols, and search for patterns. You can insert any valid GCG sequence symbol (see Appendix III) into the sequence by typing the symbol. It is inserted at the cursor.
The <Delete> key and <Ctrl>H delete the symbols to the left of the cursor, one by one.
To move the cursor to the right, use the <Right-arrow> key; to move to the left, use the <Left-arrow> key. Movements are confined to the length of the sequence.
If you type a number followed by <Return>, the cursor moves to that sequence position.
The arrow keys can be preceded by a number indicating how many symbols to move to the left or right. 10<Right-arrow> moves 10 symbols to the right.
To search for a pattern, type a / (slash) in Screen Mode. The cursor moves to the lower-left corner of the screen to let you enter a sequence pattern that you wish to find. You may type in a pattern up to 40 characters long. You can repeat the last search by simply typing /<Return>. SeqEd treats all nucleic acid sequences as circular and finds your pattern even if it wraps from the end of the sequence into the beginning. SeqEd uses the same rules for pattern definition and recognition as the programs FindPatterns, MapPlot, Map, and MapSort.
The command-line parameters -PROtein and -PERFect or the PROtein or PERFect commands in Command Mode make SeqEd treat the sequence as linear and disable the nucleic acid ambiguity meanings of the GCG sequence symbols (see Appendix III) during pattern searches.
The NUCleotide command in Command Mode tells SeqEd to recognize patterns containing IUB nucleotide ambiguity symbols during searches.
Even if SeqEd thinks your sequence is nucleotide, you can request a perfect-match search by typing = after the /. For example, /=RCT only matches RCT (case does not matter) no matter which kind of sequence SeqEd thinks you have.
You can mark a position in a sequence to which you wish to return. You give the marked position a letter (like giving it a name) using the Command Mode Mark command (see below). Then, in Screen Mode, a single quote followed by the letter used to mark the sequence moves the cursor to the position where that mark was defined.
Use <Ctrl>D to leave Screen Mode and enter Command Mode.
Here is the summary of Screen Mode commands in the on-line help:
Screen Mode
[n] is an optional numeric parameter.
G, A, T, . . . - insert a sequence character
<Delete> - delete a sequence character
<Ctrl>H - delete a sequence character
/TAACG<Return> - find the next occurrence of TAACG
(last pattern entered is the default)
1<Return> - move to start of the sequence
<Ctrl>E - move to end of the sequence
[n]<Right-arrow> - go ahead n characters
[n]<Left-arrow> - go back n characters
<Up-arrow> - go up to check sequence
<Down-arrow> - go down to original sequence
'markcharacter - go to marked position
37<Return> - go to position 37 (any positive integer)
< - go back 50 characters
> - go ahead 50 characters
<Ctrl>R - redraw the screen
<Ctrl>D - enter command mode
Use <Ctrl>D in Screen Mode to enter Command Mode. The cursor moves down to the lower-left corner of the screen next to a colon prompt after which you can enter any of the commands shown below followed by <Return>.
SeqEd command editing is modeled on OpenVMS DCL command-line editing. The <Left-arrow> and <Right-arrow> keys let you move your cursor around in a command that you have typed so you can insert or delete characters at any position. <Ctrl>E moves the cursor to the end of the line. <Ctrl>U deletes all the characters from the current cursor position to the start of the line.
SeqEd lets you modify and execute previous commands. The <Up-arrow> key displays previous commands.
If you press <Return> without entering a command, SeqEd returns to Screen Mode (described above). If you have -SINGlecommand on the command line or in your command-line initializing file, SeqEd returns to Screen Mode immediately after executing each command.
Only the capitalized portion of the commands described in the documentation below needs to be typed.
Some commands can be preceded with numeric parameters or succeeded with a file name. The square brackets ([ and ]) in the documentation below show command parameters that are optional, meaning you can leave them out.
Here is the summary of Command Mode commands you would see with the Help command:
Command Mode
Commands end with <Return>. [n] indicates an optional parameter.
s and f are numbers for start and finish of a range of interest
Only the capitalized part of the command is necessary.
EDit seqname - get a new sequence file to edit
[n] Include [seqname] - insert another sequence [at position n]
(SeqEd prompts for range and strand)
s,f Delete - delete a range of bases
[s] Check [/Blind] - check a range of bases [beginning at s]
37 - go to base 37
REDraw - redraw the screen
[n] COmment comment - insert a comment [at position n]
[n] COmment - enter comment editing mode [at position n]
[n] HEAding - edit documentary heading [at line n]
change - enter screen mode (<Return> is sufficient)
screen - enter screen mode (<Return> is sufficient)
OVERstrike - enter overstrike mode
INSert - enter insert mode
[n] Mark markcharacter - mark the sequence [at position n]
PERFect - require finds to be perfect matches
PROtein - set sequence type to PROTEIN
NUCleotide - set sequence type to NUCLEOTIDE
[s,f] Write [seqname] - write [a part of] the sequence to a file
DIGitizer - enter digitizer mode
RELoad - enter reload mode
ACCept - terminate reload mode
Help - show commands in screen and command modes
[s,f] EXit [seqname] - write [a part of] the sequence and quit
Quit - quit the editor without writing the sequence
gets a new sequence from the file you have named for editing with SeqEd. The sequence you are currently editing is lost if you have not written it out before using the EDit command.
includes another sequence within the sequence being edited at the current cursor position or at the position specified by the optional parameter. SeqEd creates two embedded comments at the start and end of the included section to show what was included. If you do not supply a file name with this command, SeqEd prompts you for one.
deletes some or all of the sequence. You must specify a beginning and ending coordinate for the range of symbols you want to delete.
lets you check a sequence entry in Screen Mode. A sequence already entered may be typed in again. If a symbol is typed that disagrees with the first entry, a ^ is printed at the point of disagreement and the terminal bell rings. While checking, the <Up-arrow> and <Down-arrow> keys move the cursor back and forth between the second entry and the original sequence, allowing you to make changes in either one as mistakes are found. If the optional starting coordinate precedes the command, it specifies where checking begins. If you wish to check your sequence without seeing the original version, type /Blind following the CHeck command (there must be a blank between the CHeck command and the /).
redraws your terminal screen. This is useful if noise in the line between your terminal and the computer has changed the screen in some unreasonable way or if a system message appears on your screen.
allows you to enter, delete or modify embedded comments to document your sequence. In its simplest use, the COmment command lets you insert new comments. You simply type the entire comment on the command line. Deletion and modification of existing comments is handled by entering Comment Mode. To do this, you type only the COmment command and optional position but no comment text. See the COMMENT MODE topic for more information.
Whenever you enter a comment, SeqEd ensures that comment-delimiting characters are placed around it. A $, <, or > must appear at each end of, and not within, your comment. (SeqEd deletes comment delimiting characters found within a comment when they are the same as the flanking comment delimiting characters.)
SeqEd inserts new comments at your current cursor position or at the position specified by the optional position number and then returns to Command Mode.
enters Heading Mode, which lets you edit the documentary heading. You can modify any part of the heading. Heading Mode is terminated with <Ctrl>D. The optional parameter specifies which line of the heading you want to start editing.
returns your session to Screen Mode. Note that the entire command is optional and a simple <Return> is equivalent.
returns your session to Screen Mode. Note that the entire command is optional and a simple <Return> is equivalent.
enters overstrike mode. Typing in a new symbol deletes the old symbol at that position and replaces it with the new symbol.
enters insert mode. Typing in a new symbol shifts all symbols from the current position to the end of the sequence by one position to the right and adds a new symbol at the current position.
You can mark a position in the sequence if you wish to return to it later. If the optional position number is absent, the position marked is the current cursor position. You give the marked position a letter (like giving it a name) using this command. Then, in Screen Mode, a single quote followed by the letter used to mark the sequence moves the cursor to the position where that mark was defined.
makes searches linear and disables the nucleic acid ambiguity meanings of the GCG sequence symbols (see Appendix III).
sets the sequence type to protein. This makes searches linear and disables the nucleic acid ambiguity meanings of the GCG sequence symbols. This also makes SeqEd ignore any set.keys file in your local directory.
sets the sequence type to nucleotide. This makes searches circular and tells SeqEd to recognize patterns containing IUB nucleotide ambiguity symbols. SeqEd also remaps the keys if a set.keys file is in your local directory.
writes the current form of the sequence into a file. If you supply starting and finishing coordinates, SeqEd only writes the indicated segment. For example, 1,56 Write would write symbols 1 to 56 into a file. If you name a file, SeqEd writes the sequence into a file with that name instead of into the input file.
enters Digitizer Mode. The lane order from left to right may be redefined. See below for a detailed discussion of digitized input.
The DIGitizer command has two options.
You can specify the left-to-right order of your lanes. The default order is alphabetic (A,C,G,T). You can change the default order with the command-line parameter -LANes=T,A,G,C or with the SeqEd command DIGitizer G,T,C,A.
Maxam and Gilbert sequencers find it useful to use -LANes=G,AG,TC,C. This order is equivalent to G,A,T,C because SeqEd uses the first of multiple letters when assigning a base to a lane. However, specifying pairs of letters causes the screen display of your gel to look more like the autoradiograph.
The second parameter with the DIGitizer command is a simple asterisk (*). This suppresses prompting for lane and menu positions. If you have been digitizing some lanes and return to the keyboard for some reason, you can go back to digitizing the same lanes without having to redefine their positions.
goes into Reload Mode, which is similar to Checking Mode, except that the reloaded sequence grows leftwards from the right end of the main sequence. This is designed to help find the overlap of two loadings of the same reaction. Mismatched bases are marked with ^ (caret) characters, as in Checking Mode. Also, you can use the arrow keys to move around in and edit either the main sequence or the reloaded sequence. When the match becomes especially good, the terminal bell rings. You are free to accept or reject SeqEd's rules of what constitutes a good overlap. See the COMMAND LINE SUMMARY topic below for more information.
terminates Reload Mode. The display of the reloaded sequence goes away, leaving you with only the main sequence with the cursor at the end, ready for more input. SeqEd helps you to decide when to ACCept an overlap, but the decision is yours.
shows the commands available in the Screen and Command Modes of SeqEd.
works exactly like Write except that your session with SeqEd ends after the sequence is written out into a new sequence file.
terminates a session with SeqEd without writing a new sequence file.
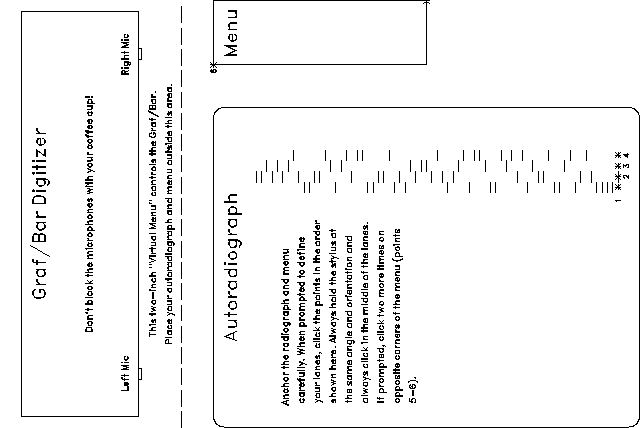
This part explains how to use the the Graf/Bar GP-7 Sonic Digitizer with SeqEd for sequence entry.
We use the Graf/Bar GP-7 Sonic Digitizer with stylus S-7 formerly manufactured by the Science Accessories Corporation which was purchased by GTCO, 7125 Riverwood Dr., Columbia, MD 21046, Phone: (410) 381-6688, Toll-free: (800) 344-4723, URL: http://www.gtco.com. Here is how to set up the GP-7 to work with SeqEd:
Remove the cover, find the DIP switches, and set the baud rate and parity according to the instructions in the Graf/Bar Operator's Manual. The parity and the baud rate must match your modem or terminal settings.
We use a special cable when connecting the Graf/Bar to the system, whether it is connected directly or through a data splitter. Build a cable between two 25-pin RS-232 connectors as follows:
Graf/Bar Computer
pin pin
1 1
7 7
3 2
We connect our Graf/Bar through a modem data splitter to the same line that we use for the terminal. The modem data splitter we use is model 232MDS from B & B Electronics, 1500 Boyce Memorial Drive, Ottawa, Illinois, 61350, USA, Phone: (815) 434-0846
The program reads input from the digitizer through a line whose logical name is DigitizerPort. This is assigned to TERM unless you (or your system manager) assign it somewhere else. If you have a port other than your terminal available, put a command like:
% name -s DigitizerPort /dev/tty11
See the figure at the back of this document to get an overview of how to lay out the digitizer.
The Graf/Bar digitizes an active area about 24 inches wide and 18 inches deep, starting 2 inches from the front of the unit. The region of interest in your radiograph must lie entirely within this active area, with the gel lanes perpendicular to the face of the digitizer. The menu must also fit in this area, with its long axis perpendicular to the digitizer. Tape or clamp the radiograph and the menu tightly so that their positions with respect to the digitizer do not change.
The digitizer controls SeqEd through a printed menu, which you place in your work area next to the radiograph. Cut out one of menus in the figures below or Fetch the file seqedmenu.fig and use the Figure program to draw more copies. The menu may be drawn to whatever absolute size you like, as long as it lies within the active area of the digitizer.
The DIGitizer command switches SeqEd into Digitizer Mode. All input comes from the digitizer stylus in this mode. After you issue this command, SeqEd asks you to define your gel lanes (described below) and to locate opposite corners of the menu. The KEYBOARD key on the menu turns off Digitizer Mode and returns control to the keyboard. A banner across the top of your screen reminds you whether the keyboard or the digitizer is in control. In Digitizer Mode, keyboard input is ignored and in Keyboard Mode, digitizer input is ignored.
The basic idea is this: click (with the stylus) in the middle of each lane where you begin (or resume) reading the gel. See the figure below. All of the points should be in the center of the lane and the stylus must always be held at the same angle and orientation. If the program is uncertain of which base to assign a digitized position, it beeps twice and asks you to redefine your lanes. The program uses the command-line value of tolerance as the basis for certainty. A tolerance of 0 is the least tolerant setting and the slightest deviation would require you to redefine your lanes. A tolerance of 1.0 is the most tolerant setting, such that any deviation is accepted. Based on our limited experience, you should not use a tolerance value less than 0.25 or greater than 0.6. The default value (0.4) was chosen because it has seldom made an incorrect assignment and does not require you to redefine the lanes too frequently. The algorithm employed is that of Staden (Nucl. Acids Res., 14, 217 (1986)).
Remember, when answering the prompts for lane position, digitize the centers of the lanes near the first bands you intend to read.
After clicking on the fourth (right-most) lane or your radiograph, you are prompted to click on two opposite corners of the menu.
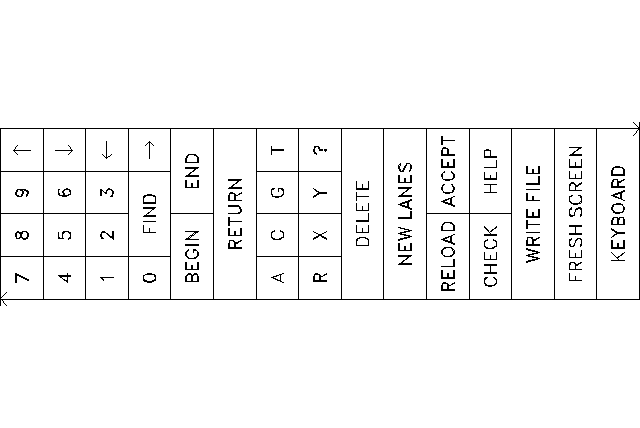
The digitizer replaces the keyboard in Digitizer Mode. You now control all of SeqEd's functions by clicking on the "keys" in the menu with the digitizer stylus. When you click on the KEYBOARD key, control is returned to the keyboard.
Enter bases into the sequence by clicking the bands on the radiograph. SeqEd behaves exactly as though you were typing uppercase letters. You can also click the menu keys labeled A, C, G and T for exactly the same effect. The menu also includes ambiguity codes R, X, and Y, which are entered into your sequence in lowercase. The question mark is like a shift key -- the next base you enter from the gel lanes after clicking the question mark is entered in lowercase.
Remove bases from the sequence by clicking the DELETE key on the menu. This behaves just like the <Delete> key, removing the base to the left of the cursor.
Move around in your sequence by clicking on the left and right arrow keys. You can click on numbers first in order to move more than one base at a time. You can click on the numbers and then click the RETURN key to move the cursor to any position. The BEGIN and END keys jump to the ends of the sequence. The FIND key lets you move to the beginning of a pattern. You can enter the pattern by clicking the letter keys on the menu ending with a click on the RETURN key.
RELOAD and ACCEPT function together to help you find the overlap of two loadings of the same sequence. RELOAD prompts you to define new lane positions. (It is not necessary to redefine the menu position.) See the documentation on RELoad and ACCept under Command Mode above.
CHECK puts you in Checking Mode so you can re-enter the data and increase your confidence. Use the up and down arrows to move between the main sequence and the check sequence.
WRITE FILE writes your sequence into a file without leaving Digitizer Mode.
FRESH SCREEN refreshes your terminal screen if line noise or broadcast messages have interrupted it.
SeqEd should handle slight bends and bulges in your lanes. However, if your bands are not being recorded correctly, it may be necessary to delete the incorrect bases and redefine your lane positions starting with a click on the NEW LANES key.
You can also use this key when you have finished one set of lanes and wish to start another without returning to Keyboard Mode.
The KEYBOARD key returns control to the terminal keyboard. You cannot use the digitizer again until you use SeqEd's DIGitizer command.
The HELP key displays the Digitizer Mode help message (below) on your terminal screen. The next click you make, whatever it is, redraws your screen the way it was before the help screen was displayed.
Digitizer Mode
37<Return> - go to position 37 (any positive integer)
<Up-arrow> - go up to check sequence (Checking Mode)
<Down-arrow> - go down to original sequence (Checking Mode)
[n]<Right-arrow> - go ahead [n] characters
[n]<Left-arrow> - go back [n] characters
FIND TAACG<Return> - find next occurrence of TAACG
Begin - go to beginning of sequence
End - go to end of sequence
A C G T R X Y - insert A C G ... into sequence (at cursor)
? - enter next sequence character in lowercase
<Delete> - delete a sequence character
New Lanes - define new lanes (another loading) for this
fragment
Reload - define new lanes and slide new sequence
back over old until overlap is "Accepted"
Accept - accept overlap from "Reload"
Check - enter Checking Mode to re-enter and verify
sequence
Write File - write out fragment into a file
Fresh Screen - redraw the terminal screen
Keyboard - return control to terminal keyboard
Click the digitizer anywhere to go back to SEQED:
Comment Mode allows you to add, change, or delete embedded comments and helps you move quickly to any position in your sequence where a comment is associated. To enter Comment Mode, you must first enter Command Mode with <Ctrl>D.
If you type the COmment command without any comment text, SeqEd creates a new, empty comment at the position indicated by the optional sequence position number, if present, or at your current position in the sequence. The cursor moves to the part of the screen where embedded comments are displayed. Initially, the cursor is adjacent to a position number followed by an empty comment. You may then type a new comment or move to an existing comment that you wish to modify. Only one new comment can be created each time you enter Comment Mode.
While in Comment Mode you can move around in the comment using the <Left-arrow> and <Right-arrow> keys, insert text by typing, or delete text using the <Delete> key or <Ctrl>H. <Ctrl>E positions the cursor at the end of the comment. <Ctrl>U deletes all characters from the beginning of the comment to the cursor position. You can move from one comment to another using the <Up-arrow> or <Down-arrow> keys.
When you move the cursor off of a comment that is empty, the comment is deleted. You can delete a comment by entering Comment Mode, moving to the end of the comment you wish to delete, and using <Ctrl>U. When you move to another comment or leave Comment Mode, the comment disappears. Likewise, the empty comment created when you enter Comment Mode is deleted if you don't type anything at the new comment position.
Comments must start and end with one of the characters <, >, or $. A comment must start and end with the same delimiting character. If you try to move your cursor off of a comment that does not have one of these characters at the ends, or if the delimiters aren't identical, then SeqEd makes sure the delimiters are corrected.
As you move to different comments, your position in the sequence in Screen Mode changes to the symbol with which that comment is associated. This allows you to move quickly to any symbol with which a comment is associated when you leave Comment Mode. By marking your place with a comment at the end of one session with SeqEd, you can easily restore your place at the next session.
To exit Comment Mode, press <Return> or use <Ctrl>D.
Comments may be associated with any base. They stay with that base, even though the base's position may change, unless the base is deleted (see below). They can also be associated with either end of the sequence. For example, you may issue the command, 0 CO to create a comment associated with the left end of the sequence. This comment must be delimited with < (SeqEd makes sure of this). Similarly, a comment can be created at the extreme right of the sequence and must be delimited with > or $.
Comments can document a whole fragment as well as an individual sequence symbol. For example, the Include command automatically puts an identifying comment at each end of the included fragment. The characters < and > were selected as comment delimiters because they imply direction; the comments bracketing the included fragment point at the fragment. A >-comment is associated with the first base of the fragment and a <-comment with the last. When the sequence is saved in a file, all >- and $-comments are written before the base they are associated with and all the <-comments after. This way the bracketing comments surround the entire fragment and point to it.
Between two bases in a sequence file there may be several comments. The <-comments are always associated with the base to the left, the >- and $-comments with the base to the right.
The only way to delete a comment is to go into Comment Mode and delete all the characters of the comment. When you move your cursor away from the empty comment, it goes away.
If you delete a base with which a comment is associated, the comments do not go away. They just attach themselves to adjacent bases. To preserve the properties of fragment bracketing comments, the <-comments become associated with the left-hand base, the >- and $-comments with the right-hand base.
Heading Mode allows you to edit the documentation that appears above the sequence. When a new sequence is edited, SeqEd goes directly into Heading Mode to let you identify the new sequence.
SeqEd lets you enter Heading Mode by using the HEAding command.
Use <Ctrl>D to return to Command Mode.
You can move around using the arrow keys and make insertions and deletions as you wish. Although the editing window is only four lines high, it scrolls over the heading vertically to let you see and modify any part. <Ctrl>E positions the cursor at the end of the current line.
As with many text editors, typing inserts text at the cursor and the <Delete> key or <Ctrl>H delete characters to the left of the cursor. <Ctrl>U deletes everything from the current cursor position to the start of the line; <Return> creates a new line starting at the current position in the heading.
While you are editing a sequence, SeqEd records your session in a file called seqed.log. This file is automatically deleted when the editor exits normally. If you are accidentally disconnected or the system crashes, your work can be recovered by logging back in, moving to the directory where the crash occurred, and running SeqEd again. SeqEd finds seqed.log and restores the sequence to the state it was in just before you were cut off.
If you do not want SeqEd to restore the session, delete the file seqed.log.
The total length of all vector sequences specified with the -VECtors command-line parameter may not exceed 100,000 bases. If the total vector sequence length exceeds 100,000 bases, SeqEd notifies you that only the first 100,000 vector bases will be checked.
SeqEd only works on terminals that can provide screen support. Your system manager may be able to help if your terminal is not behaving correctly.
Many terminal conditions must be set if you are using the Graf/Bar sonic digitizer; see the paragraph on Preparation under the DIGITIZER MODE topic.
SeqEd was originally designed by Paul Haeberli and implemented for VAX/VMS by Paul Haeberli and John Devereux. It was completely revised for GCG Version 4 by William Winsborough. The digitizer interface and the RELoad command were implemented for Version 5 by Philip Delaquess. We are very grateful for the collaboration of Dr. William Boorstein.
When it opens a new sequence file, SeqEd initially assumes it is nucleic acid. When you write the file, SeqEd examines the sequence to see if it contains only IUB-IUPAC nucleotide symbols in the first 300 symbols. If so, it writes the new sequence as a nucleic acid sequence; if not, it writes it as a peptide sequence.
When it opens a pre-existing GCG sequence file, SeqEd obtains the sequence type (nucleotide or protein) from the Type: field of the dividing line (the line that contains two successive periods). If the Type: field is absent, as in the case of sequence files created prior to Version 7 of the Wisconsin Package, SeqEd infers the type of the sequence from the composition of the sequence characters. When SeqEd writes the edited file, it writes the Type: field according to its current understanding of the sequence type.
It is possible for SeqEd to make a mistake. If the Type: field of an existing file is incorrect, SeqEd will accept the incorrect type; it doesn't check the composition in this case. For files without a Type: field, it is possible for SeqEd to infer the wrong sequence type. For example, a peptide sequence that contains only those amino acids that share IUB-IUPAC symbols with nucleotides will be incorrectly typed as nucleic acid (see Appendix III).
You can override SeqEd's assignment of sequence type in two ways. When you run SeqEd, you can add -PROtein or -NUCleotide to the command line to tell SeqEd which type of sequence it will be editing. Once SeqEd is running, you can use the Command Mode commands PROtein and NUCleotide to force the assignment of sequence type.
All parameters for this program may be added to the command line. Use -CHEck to view the summary below and to specify parameters before the program executes. In the summary below, the capitalized letters in the parameter names are the letters that you must type in order to use the parameter. Square brackets ([ and ]) enclose parameter values that are optional. For more information, see "Using Program Parameters" in Chapter 3, Using Programs in the User's Guide.
Minimal Syntax: % seqed [-INfile1=]sample.seq
Prompted Parameters: None
Local Data Files:
set.keys (must be in your current working directory to be used)
Optional Parameters:
-SINGlecommand automatically returns to screen mode after commands
-PROtein sets sequence type to protein, and sets find to
search for perfect symbol matches
-NUCleotide sets sequence type to nucleotide, and sets find to
allow nucleotide ambiguity symbol matches
-PERFect sets find to search for perfect symbol matches,
even if sequence type is nucleotide
-VECtors=GB:SynpBR322 highlights sequences from pBR322
-SITes=GAATTC highlight GAATTC patterns
-LANes=A,C,G,T sets the default lane order for digitizer
-MINOverlap=10 sets minimum overlap length for Reload command
-PCTOverlap=95 sets stringency for the Reload command
-TOLerance=0.4 sets tolerance for digitizing ambiguity (0 to 1),
with 1 being the most tolerant
The files described below supply auxiliary data to this program. The program automatically reads them from a public data directory unless you either 1) have a data file with exactly the same name in your current working directory; or 2) name a file on the command line with an expression like -DATa1=myfile.dat. For more information see Chapter 4, Using Data Files in the User's Guide.
You can use the program SetKeys to create a set.keys file that tells the SeqEd, GelEnter, LineUp,GelAssemble, and SeqLab sequence editors how to interpret the letters you type at the terminal. When entering gel readings, it is useful to have the symbols for G, A, T, and C under the fingers of one hand in the same positions as the lanes in your gel. SeqEd, GelEnter, LineUp, GelAssemble, and the SeqLab sequence editor automatically read the file set.keys if it is present in your local directory. If set.keys is absent, or if the sequence type is set to Protein (in SeqEd and LineUp only) the terminal keys retain their conventional meanings.
If you have a set.keys file in your directory, SeqEd, GelEnter, LineUp, and GelAssemble only respond to the keys that it redefines. You can edit the file set.keys with a text editor if some of the keys you want to use are not in it. Any keys not mentioned in set.keys appear to be dead in these sequence editors. In the SeqLab sequence editor, keys that are not redefined retain their normal meanings.
Several keys are vital for the control of SeqEd, LineUp, GelEnter, and GelAssemble; this means you are not allowed to redefine the keys for /, [, ], {, }, (, ), :, ,, 1, 2, 3, 4, 5, 6, 7, 8, 9, 0, <Ctrl>R, <Ctrl>D, <Ctrl>H, <Return>, and <Ctrl>E.
You can set the parameters listed below from the command line. For more information, see "Using Program Parameters" in Chapter 3, Using Programs in the User's Guide.
sets SeqEd to return automatically to Screen Mode after every command in Command Mode.
sets the sequence type to be protein, and makes pattern searches use perfect symbol matches. SeqEd treats protein sequences as linear and will not find patterns that start at the end and continue into the beginning of the sequence. Furthermore, -PROtein causes SeqEd to ignore any set.keys file in your local directory.
sets the sequence type to be nucleotide, and makes pattern searches use nucleotide ambiguity symbol matches (unless you force the program to use perfect symbol matches by including -PERFect on the command line or by entering the PERFect command in Command Mode.) SeqEd treats nucleotide sequences as circular and will find patterns that start at the end and continue into the beginning of the sequence. Furthermore, -NUCleotide causes SeqEd to use a set.keys file in your local directory.
makes pattern searches use perfect symbol matches. Normally if you type /GARC in Screen Mode, the patterns GAAC or GAGC could be found. If you have -PERFect on the command line, /GARC would only find the pattern GARC. This also makes SeqEd treat sequences as linear and not find patterns that start at the end and continue into the beginning of the sequence.
tells SeqEd which cloning vector or vectors are of interest to you. SeqEd checks your sequence against them to make sure you are not entering a vector sequence. If it finds that you are entering vector sequence, the terminal bell rings and the vector sequence characters are highlighted with reverse video.
tells SeqEd to highlight enzyme recognition sites that interest you.
establishes the default left-to-right order of gel lanes. The default may be over-ridden when you issue a DIGitizer command in Command Mode.
sets the minimum overlap length regarded as meaningful by the RELoad command. SeqEd ignores matches shorter than this, even if they are perfect. However, you are always free to end a reload with the ACCept command.
sets the minimum percentage of matching bases regarded as meaningful by the RELoad command. In Reload Mode, when the overlap is long enough and good enough, the terminal bell rings to alert you. Again, you have complete freedom to reject or ACCept SeqEd's opinion.
sets the tolerance for digitizing. When digitizing, the program must determine which base lane the sonic pen has touched. Since the gel lane may bend, the program must have some tolerance for deviation. The tolerance value determines how great this deviation can be before you must redefine your lanes. A tolerance of 0 is the least tolerant setting and the slightest deviation would require you to redefine your lanes. A tolerance of 1.0 is the most tolerant setting such that any deviation is accepted. Based on our limited experience, you should not use a tolerance value less than 0.25 or greater than 0.6. The default value (0.4) was chosen because it has seldom made an incorrect assignment and does not require you to redefine the lanes too frequently. The algorithm employed is that of Staden (Nucl. Acids Res., 14; 217 (1986)).
[ Program Manual | User's Guide | Data Files | Databases ]
Documentation Comments: doc-comments@gcg.com
Technical Support: help@gcg.com
Copyright (c) 1982, 1983, 1985, 1986, 1987, 1989, 1991, 1994, 1995, 1996, 1997, 1998 Genetics Computer Group Inc., a wholly owned subsidiary of Oxford Molecular Group, Inc. All rights reserved.
Licenses and Trademarks Wisconsin Package is a trademark of Genetics Computer Group, Inc. GCG and the GCG logo are registered trademarks of Genetics Computer Group, Inc.
All other product names mentioned in this documentation may be trademarks, and if so, are trademarks or registered trademarks of their respective holders and are used in this documentation for identification purposes only.