Title
Population-Based Structural Connectome Analysis
Introduction
Many challenging issues, such as statistical variability in a population, arise from the study of structural connectome maps by using diffusion MRI tractography data. Addressing these challenges requires the development of fast and reliable approaches for processing high-dimensional diffusion data from hundreds (or even thousands) of subjects. We aim to develop a reliable Population-based Structural Connectome (PSC) Mapping framework to construct population structural connectome maps on a common space (or template) while accounting for individual variabilities. Our goal is to provide a set of tools for researchers to conveniently explore structural connectivity. These tools will allow one to construct and visualize individual structural connectome in different forms, from coarse to detail, e.g. (1) binary network, (2) weighted network, (3) tensor network and (4) streamline-based connnectome. At the binary network level, the connection two regions of interest are represented as 0 (not connected) or 1 (connected). At the weighted network level, a scalar number is assigned to indicate the "connection strength" of these two regions. At the tensor network level, a set of scalar numbers is assigned to characterize this connection. Eventually, at the streamline level, the connection is characterized by the reconstructed fibers, which gives the most detailed information about this connection.Questions
There are two key questions we try to answer: (1) How to reliably construct the connectome for population-based analysis? (2) How to efficiently summarize the data to allow meaningful statistical analysis for a population of individuals?Methods
The whole pipeline includes five different modules as illustrated in Figure 1. Module (1) and (2) use state-of-the-art tractography algorithm to reconstruct the fiber curves using dMRI and structural MRI. Module (3), (4) and (5) are newly developed procedures for reliably extract each connection and efficiently represent each connection. The outputs of this pipeline are: binary networks, weigthed networks, tensor networks and streamline connectivity matrices.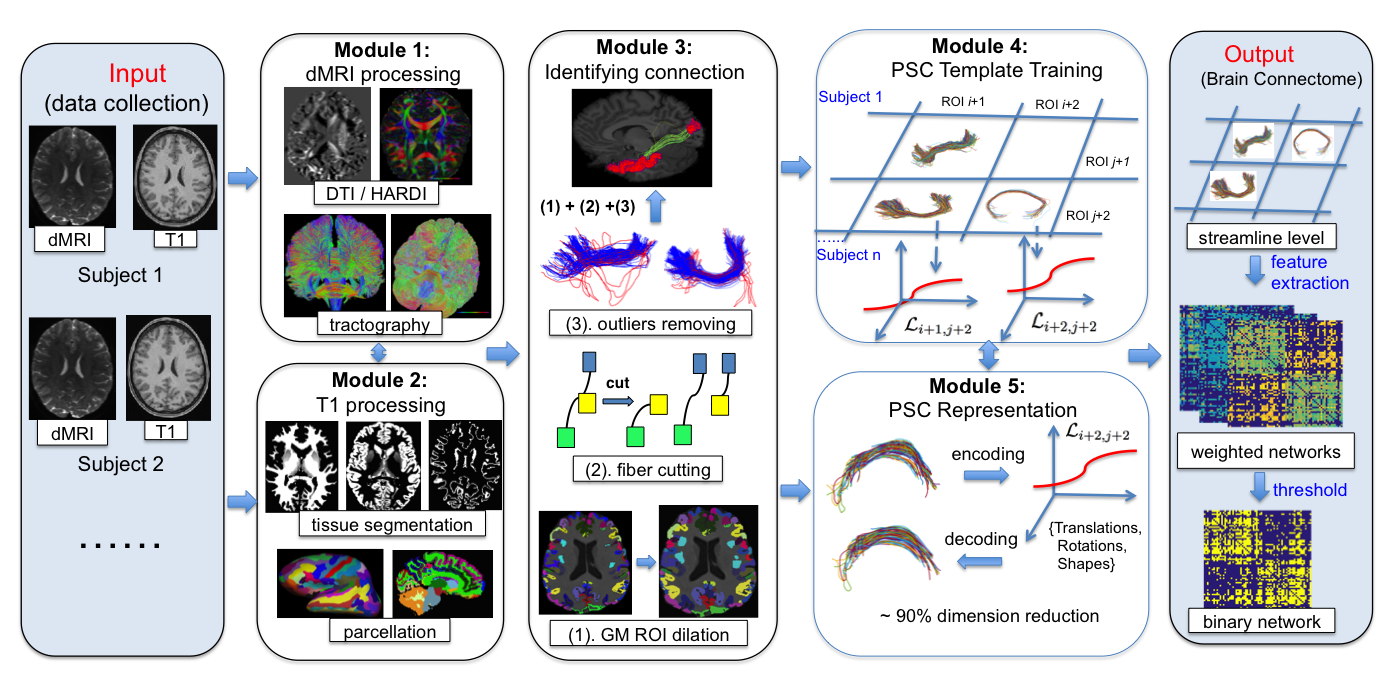
Findings
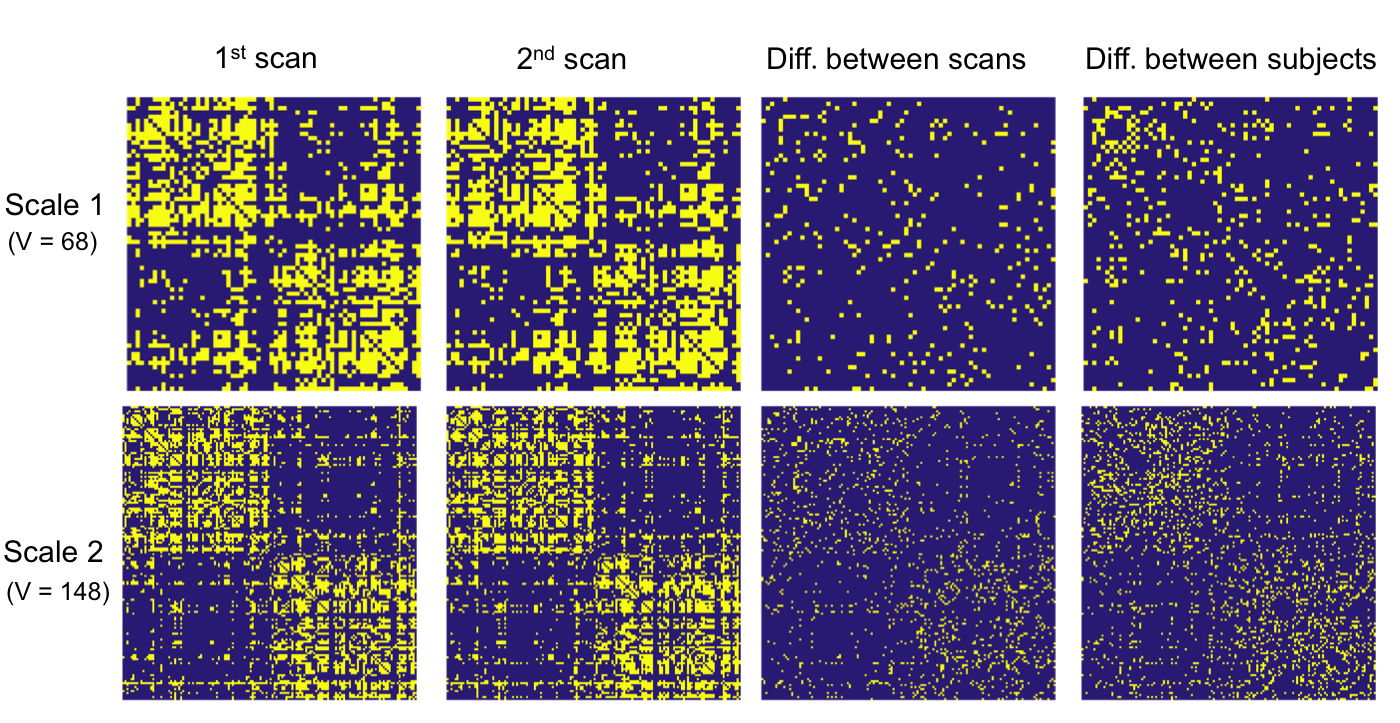
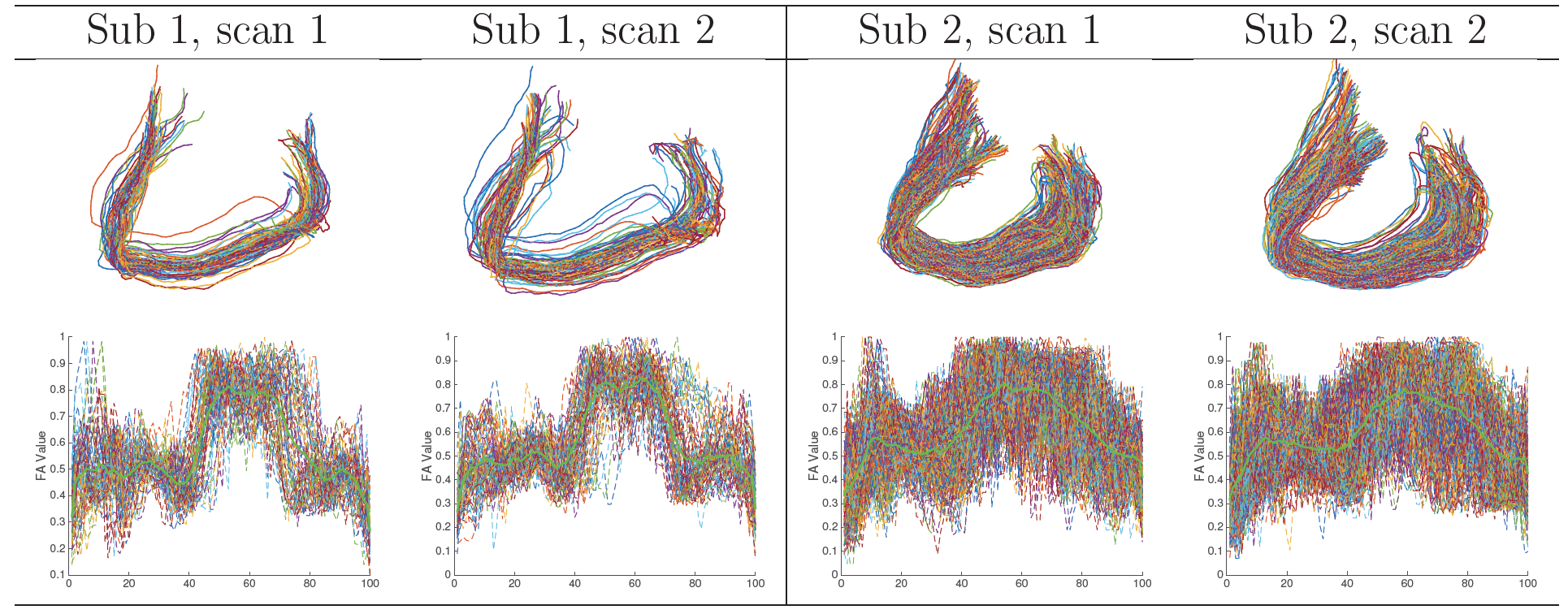
The developed PSC allows one to perform population based structural connectivity analysis. Figure 2 shows the binary and weighted network extracted from PSC.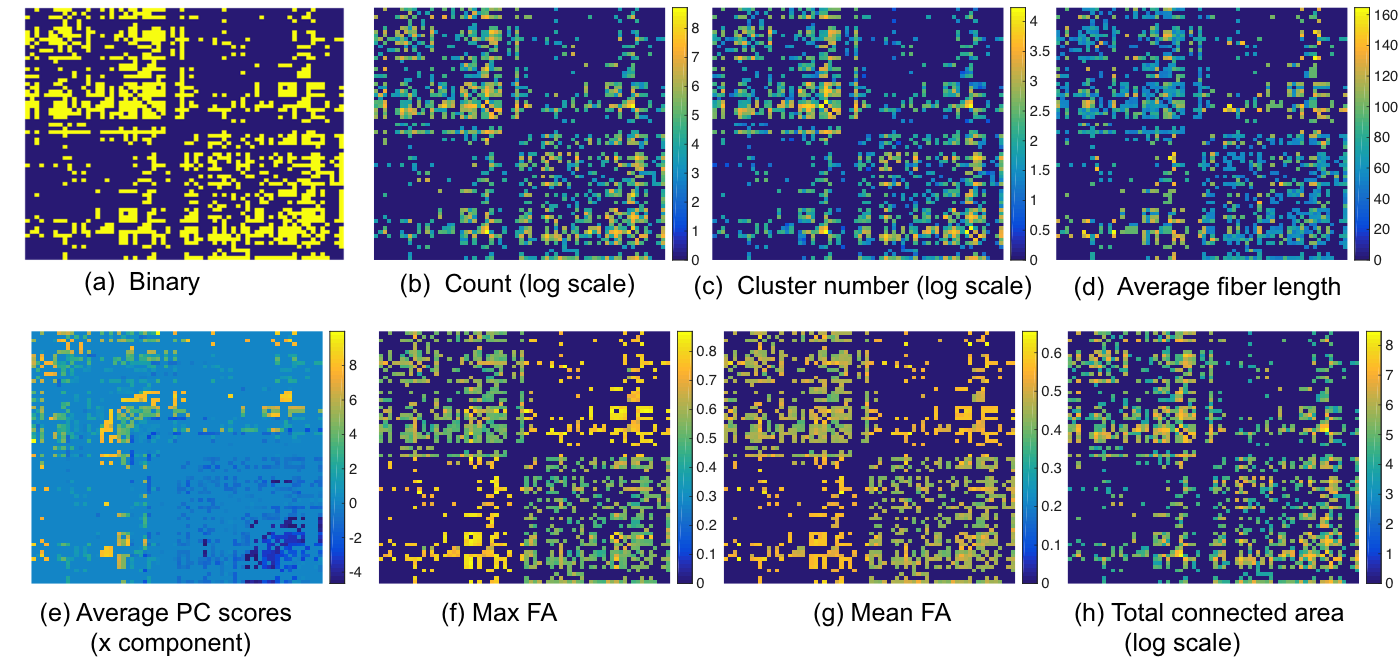 Reproducibility: we dedicate to extract robust and reliable connectivity from the dMRI. Figure 3 shows two examples of the binary network matrices obtained from two different scans of the same person. Column 3 shows the difference between two scans in column 1 and 2 (for the same subject). Column 4 shows the difference between network matrices extracted from two randomly selected persons (for different subjects). Figure 4 shows the streamlines connecting part of left and right frontal sulci and these streamlines are constructed from four scans of two subjects. The second row shows the FA values along each streamline; the bold green line in each plot is the mean of these FA curves.
Reproducibility: we dedicate to extract robust and reliable connectivity from the dMRI. Figure 3 shows two examples of the binary network matrices obtained from two different scans of the same person. Column 3 shows the difference between two scans in column 1 and 2 (for the same subject). Column 4 shows the difference between network matrices extracted from two randomly selected persons (for different subjects). Figure 4 shows the streamlines connecting part of left and right frontal sulci and these streamlines are constructed from four scans of two subjects. The second row shows the FA values along each streamline; the bold green line in each plot is the mean of these FA curves.
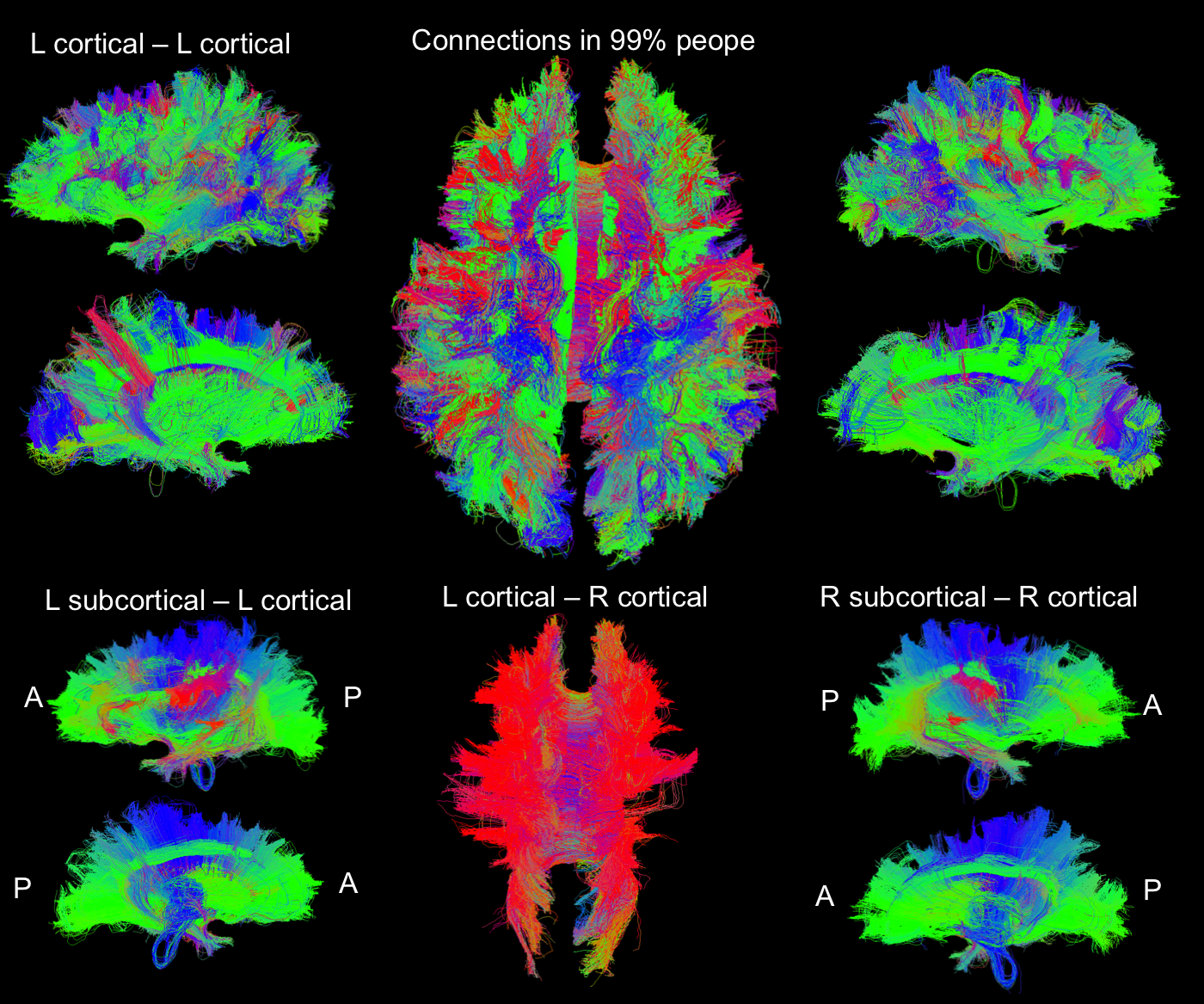
 Common Structural Connection: We are interested in finding the share connections among a population of individuals. We processed ~900 subjects in HCP dataset and extracted the connections that are shared by 99% of people in this set. Figure 5 shows the common connections in a randomly selected subject.
Common Structural Connection: We are interested in finding the share connections among a population of individuals. We processed ~900 subjects in HCP dataset and extracted the connections that are shared by 99% of people in this set. Figure 5 shows the common connections in a randomly selected subject.