My research is motivated by large-scale complex data sets from recent genomic and familial studies and by important biological questions that emerge from the analysis of these data. My current interests can be divided into two parts: 1) Development of methods and software for the accurate measurement of high-throughput genomic data; 2) Development of software and validation of statistical approaches for personalized cancer risk prediction (see publications and software for details).
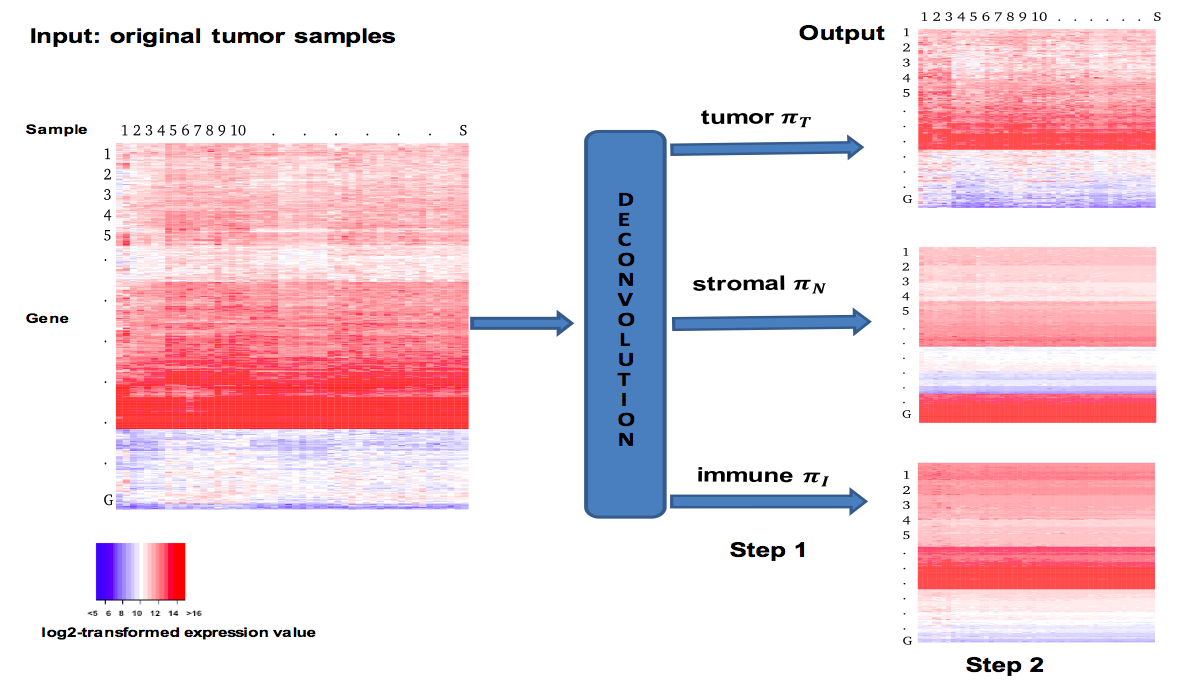
1). It is non-trivial to extract genomic information of interest from the raw signals that come directly from chemical or physical reactions. Current high-throughput technologies have all inevitably incorporated multi-level confounders that affect these observed signals. The large amount of data they produce also make it difficult to calibrate these technologies using "gold standards", usually generated by experiments that are more accurate but are low-throughput and expensive. In this area, my work is focused on the accurate interpretation of raw high-throughput signals using statistical modeling. I have worked with high-throughput data measuring genomes such as copy number and single nucleotide variants, as well as those measuring transcriptomes such as expression and alternative splicing. Currently, my research group is focusing on integrating multi-platform information in order to better understand complex human diseases. In particular, we are looking in to non-small cell lung cancer (NSCLC) through collaboration with clinicians and pathologists at the University of Texas MD Anderson Cancer Center.
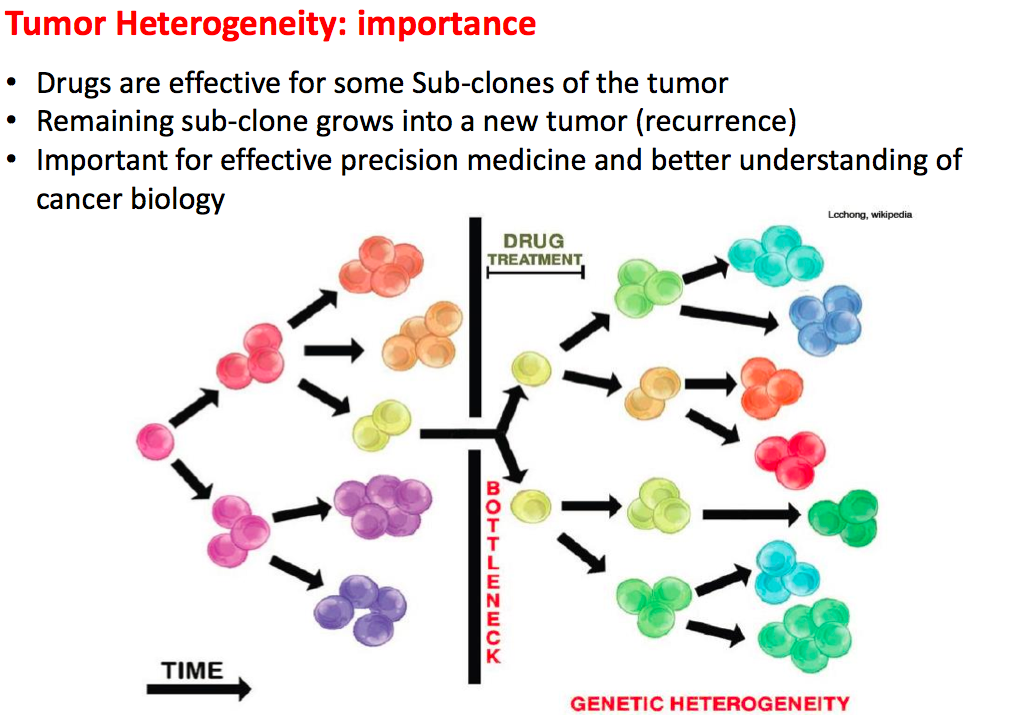
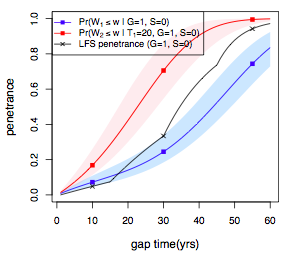
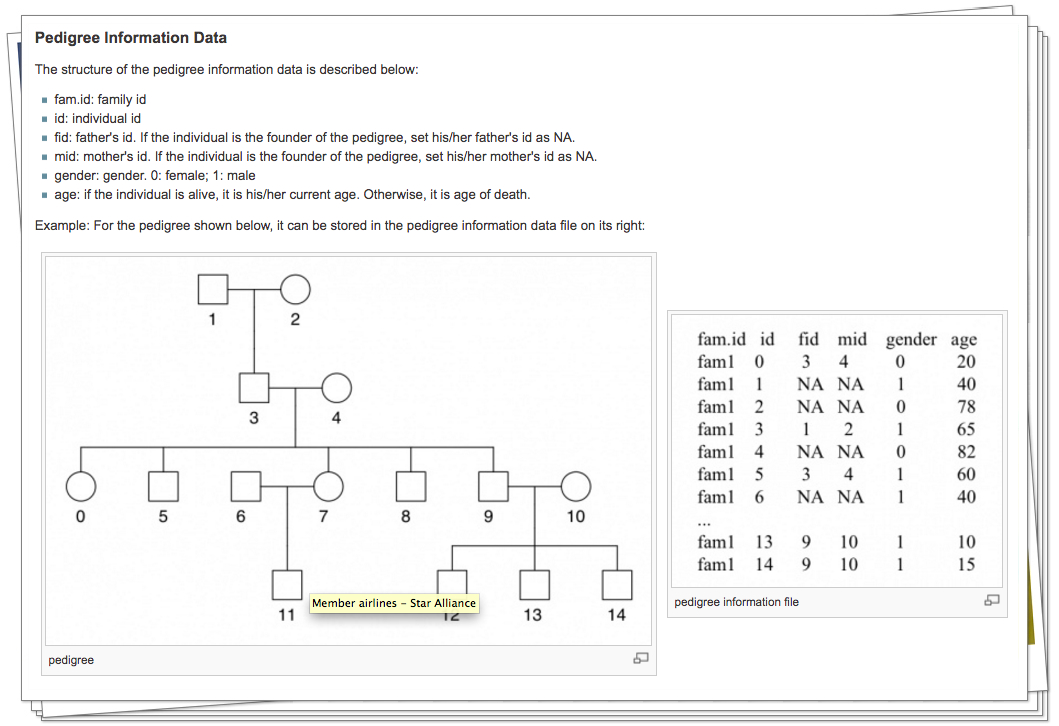
2). Cancer results from the accumulation of multiple genetic mutations. Germline mutations in specific genes predisposes the carrier to the development of cancer; the increased risk is also known as "inherited susceptibility". This inheritance results in familial clustering of cancers, known as "familial cancer syndromes". Clinical researchers utilize model-based prediction algorithms to identify mutation-carrying cancer patients at earlier and more treatable stages and/or to identify healthy individuals at high risk of developing cancer in future. As a result, Mendelian carrier probability models are based on Bayesian methods using detailed family history as input, and have shown performances better than empirical models using regression or classification trees alone. In this area, my work is focused on a) applying Mendelian models to cancers of interest for personalized risk assessment and b) developing methodologies for evaluation of risk assessment models using family and correlated data.