Statistical Bioinformatics Lab
Dr. Wenyi Wang is a Professor of Bioinformatics and Computational Biology and Biostatistics at the University of Texas MD Anderson Cancer Center. She received her PhD from Johns Hopkins University and performed postdoctoral training in statistical genomics at UC Berkeley with Terry Speed and genome technology at Stanford with Ron Davis. Wenyi’s research includes significant contributions to statistical bioinformatics in cancer, including MuSE for subclonal mutation calling, DeMixT for transcriptomic deconvolution. More recently, she led a pan-cancer characterization of genetic intra-tumor heterogeneity in subclonal selection, as well as a pan-cancer biomarker identification through integrative deconvolution of transcriptomic/genomic data. Her group is focused on the development of computational methods to study the evolution of cancer cells, and further develop risk prediction models to accelerate the translation of biological findings to clinical practice.
Pre-doctoral and post-doctoral fellow positions are available (see the biostatistics position and the cancer genomics position). Please inquire with Dr. Wang.

Current Research Directions
Deconvolution and single-cell modeling for intra- and inter- tumor heterogeneity
Tissues contain diverse cell types, each defined by unique transcriptional patterns that can be studied through RNA expression data. This also applies to tumors, where transcriptomics offers insights into cancer. Single-cell RNA sequencing (scRNA-seq) provides detailed data but is often costly and challenging for large-scale use. Bulk RNA-seq is a more affordable alternative, though it mixes signals from different cell types. To address this, deconvolution methods like [DeMixSC] separate these signals, enabling better analysis of cell proportions and disease mechanisms. In cancer research, deconvolution helps differentiate tumor from non-tumor cells, providing insights into pathways, prognosis, and heterogeneity [DeMixT, TmS]. Spatial transcriptomics TmS builds on this by adding another dimension, preserving the spatial arrangement of cells to help map tumor microenvironments (TME). This spatial context provides crucial insights into how cells interact within their environments, which is essential for understanding tumor progression. Meanwhile, the use of foundation models is revolutionizing the field by integrating bulk, single-cell, and spatial data, leading to more comprehensive analyses and deeper insights. These advancements pave the way for more effective and personalized treatment strategies.
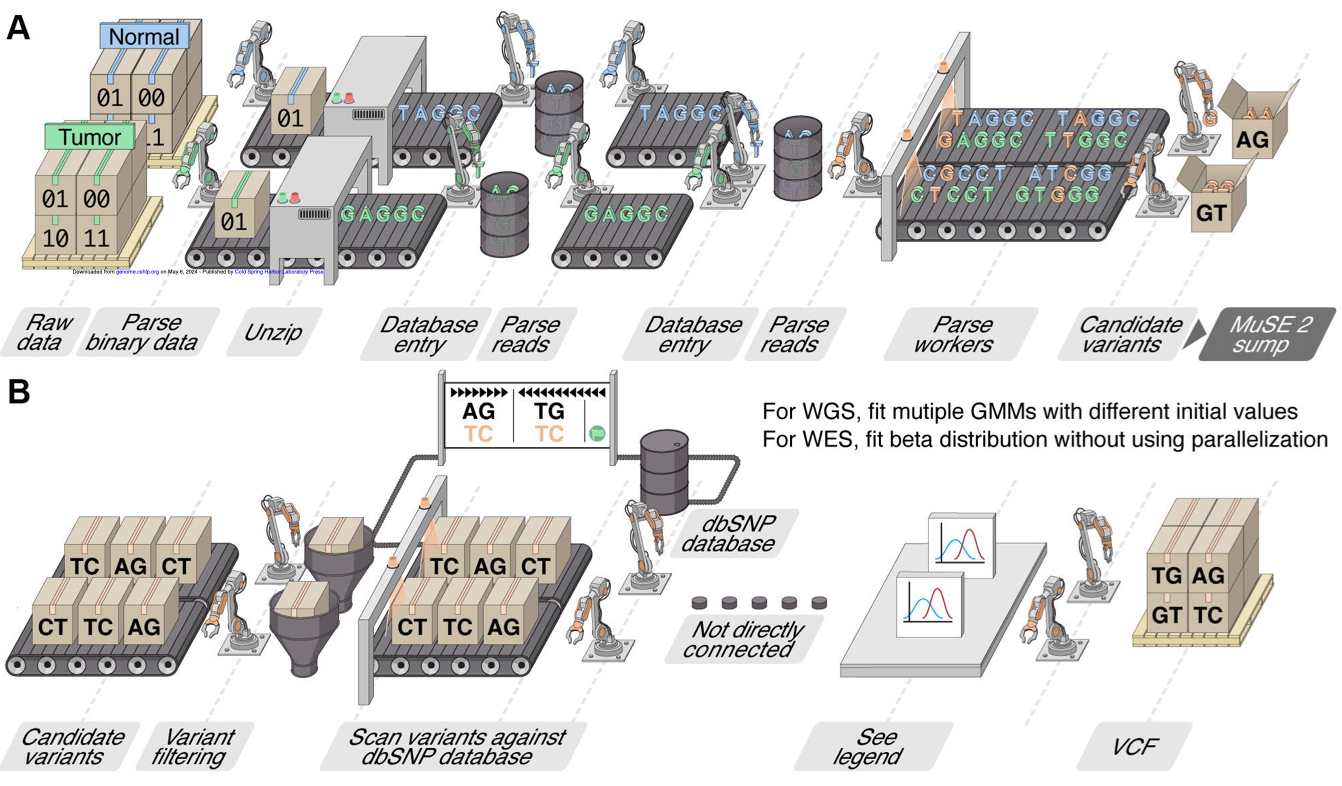
Mutation calling and subclonal reconstruction
Cancer is driven by genetic mutations, including single nucleotide variations (SNV), copy number alterations (CNA), and structural variations (SV), which influence tumor behavior, such as growth rate, treatment resistance, and metastasis. Identifying these mutations is critical for cancer research. Whole-genome sequencing (WGS) and whole-exome sequencing (WES) are key tools, but many current mutation-calling methods are slow, limiting large-scale analysis. My lab is addressing this with [MuSE2], a faster, more efficient mutation calling method that facilitates large dataset analysis and advances precision medicine. My lab is also interested in improving methods for reconstructing subclonal structures, which which are critical for understanding treatment resistance. Genetic variation within subclonal populations can lead to treatment resistance and improved fitness for cancer cells and improved fitness to cancer cell populations. Our effort on developing software tools like [CliPP] addresses limitations in previous methods, such as the lack of intra-tumoral heterogeneity characterization across cancer types and the reliance on extensive computational resources or prior knowledge [Characterizing ITH]. These advancements provide deeper insights into cancer evolution, helping clinicians improve patient outcomes.
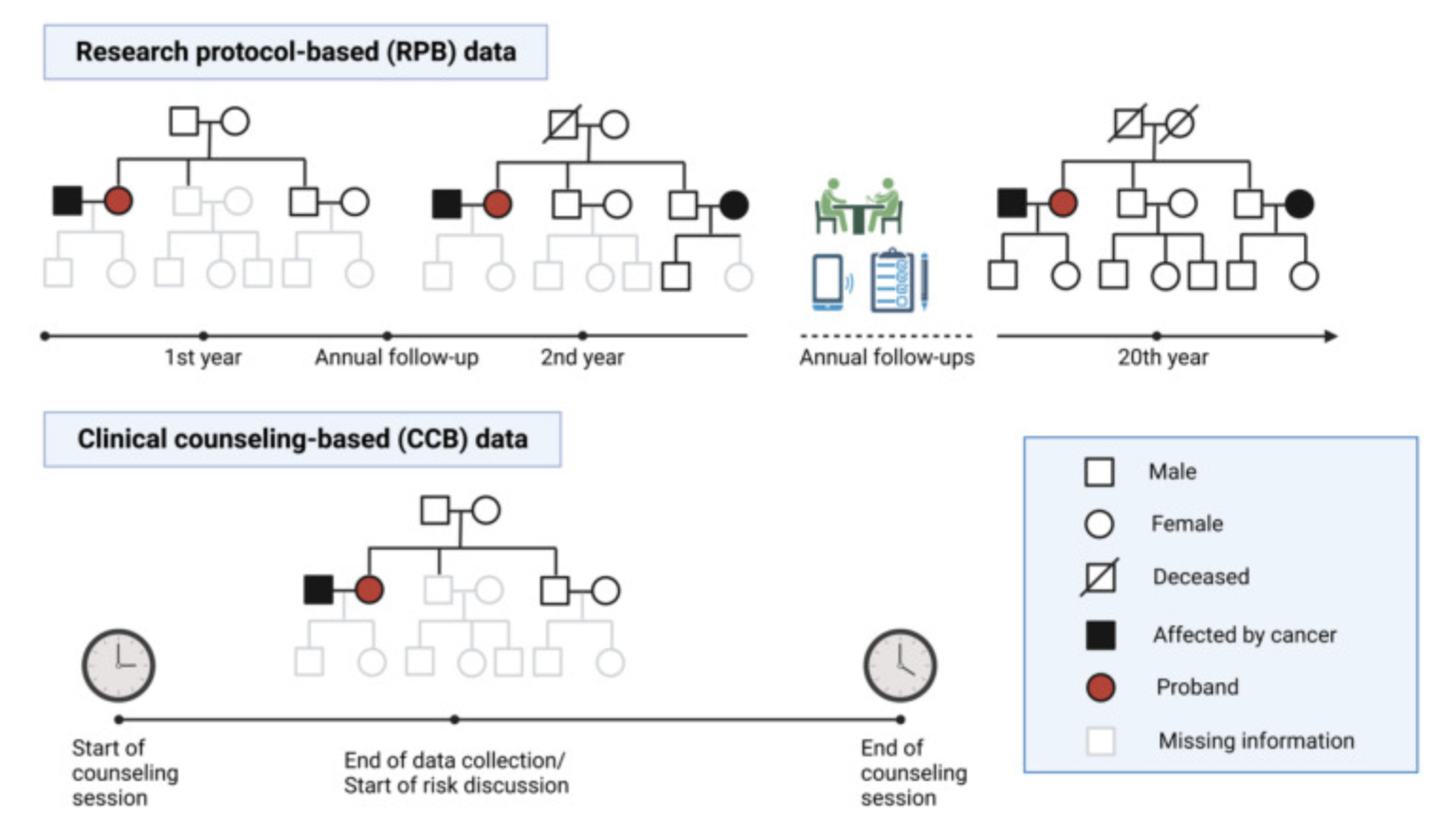
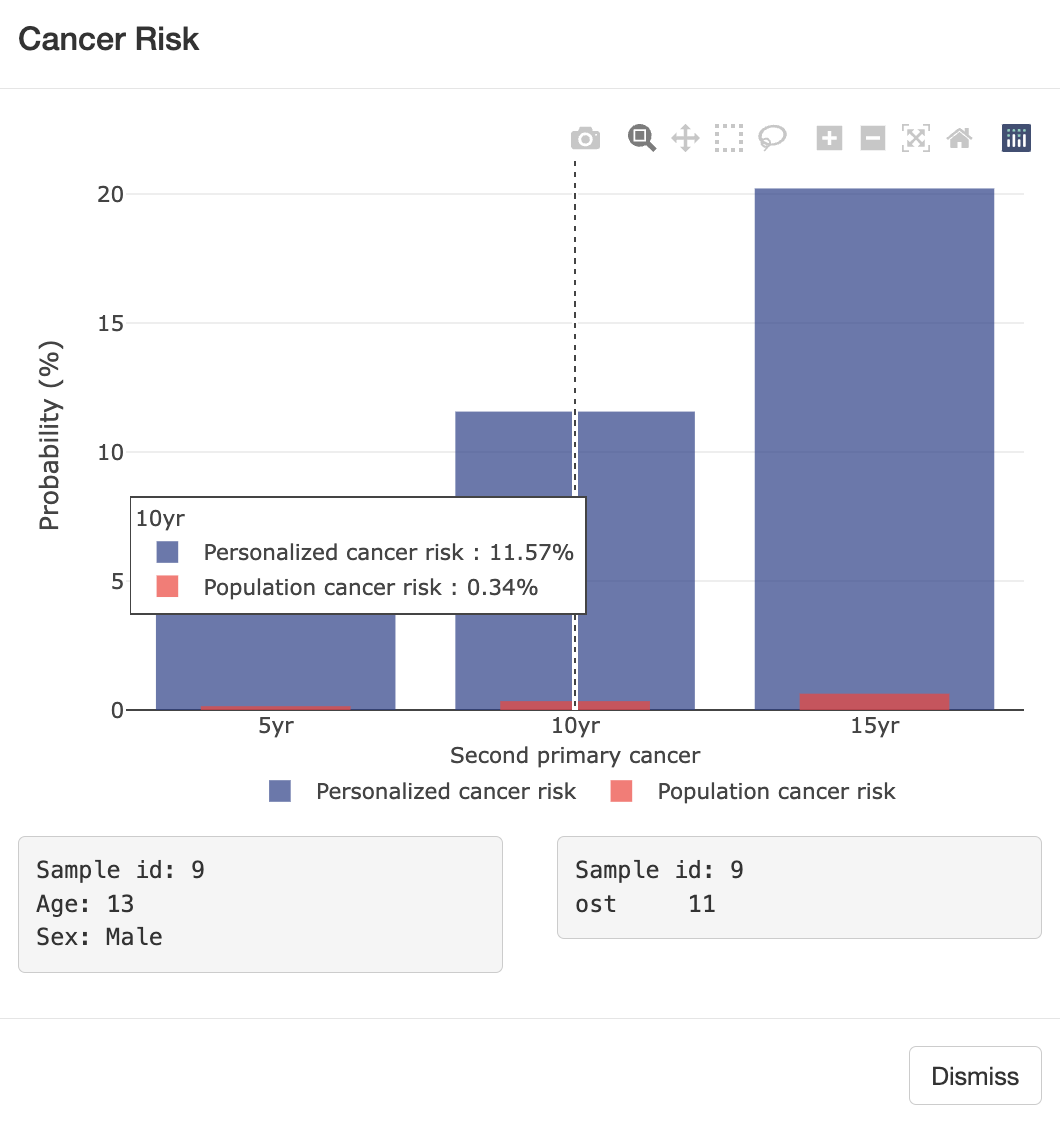
Semi-parametric survival modeling for cancer risk prediction
Cancer survivors represent a fast-growing yet under-studied population with respect to cancer risk, particularly for second primary cancers, which frequently occur in survivors of breast and bladder cancer. Current risk assessments often overlook prior cancers due to limitations in large databases like SEER, which mainly account for age and sex. To address this, my lab studies patients with Li-Fraumeni syndrome (LFS), a hereditary condition linked to higher cancer risk. LFS patients often develop multiple primary cancers, offering a unique opportunity to study cancer risk while accounting for additional factors like mutation status. Using LFS data, we developed [LFSPRO] to predict both first and second primary tumors in LFS families. These insights can help physicians and genetic counselors provide personalized treatment and screening plans, aiming for early detection of cancers in survivors and LFS patients [Personalized Risk Prediction].
PI: Wenyi Wang
Department of Bioinformatics and Computational Biology
Wenyi Wang (王文漪), Professor, Department of Bioinformatics and Computational Biology, Division of Basic Science Research, The University of Texas MD Anderson, Cancer Center, Houston, Texas
Curriculum Vitae