Statistical Bioinformatics Lab
Cancer is a complex disease shaped by layers of genetic and transcriptomic heterogeneity. The Wang laboratory is dedicated to advancing statistical bioinformatics to unravel this complexity. We develop computational frameworks that uncover the dynamics of tumor evolution and tumor microenvironment (i.e., cell-type specific transcriptional activities), and clonal architecture across diverse cancer types. We developed tools such as MuSE to enable fast and accurate mutation calling, or DeMixT to provide robust tumor-specific transcriptome deconvolution. Additionally, with over 15 years of experience in cancer risk modeling, we utilize Bayesian statistics and machine learning to develop software tools for clinical cancer prevention and prognosis. Our current research directions are 1) multi-omic deconvolution to study DNA–RNA dynamics in cancer, and 2) cancer risk modeling using machine learning and Bayesian models. In collaboration with clinicians and experimental biologists, we translate these insights into testable hypotheses and clinically meaningful advances. We are equally committed to fostering a research community where statistical rigor and artificial intelligence come together to push the boundaries of cancer discovery.
Pre-doctoral and post-doctoral fellow positions are available (see the cancer genomics position). Please inquire with Dr. Wang.

Current Research Directions
Multi-omic deconvolution to study DNA–RNA dynamics in cancer
Cancer is driven by genetic mutations, including single nucleotide variations (SNV), copy
number alterations (CNA), and structural variations (SV), which influence tumor behavior,
such as growth rate, treatment resistance, and metastasis. Identifying these mutations is
critical for cancer research. While whole-genome sequencing (WGS) and whole-exome sequencing
(WES) are key tools, basic steps like somatic mutation calling can be slow, limiting
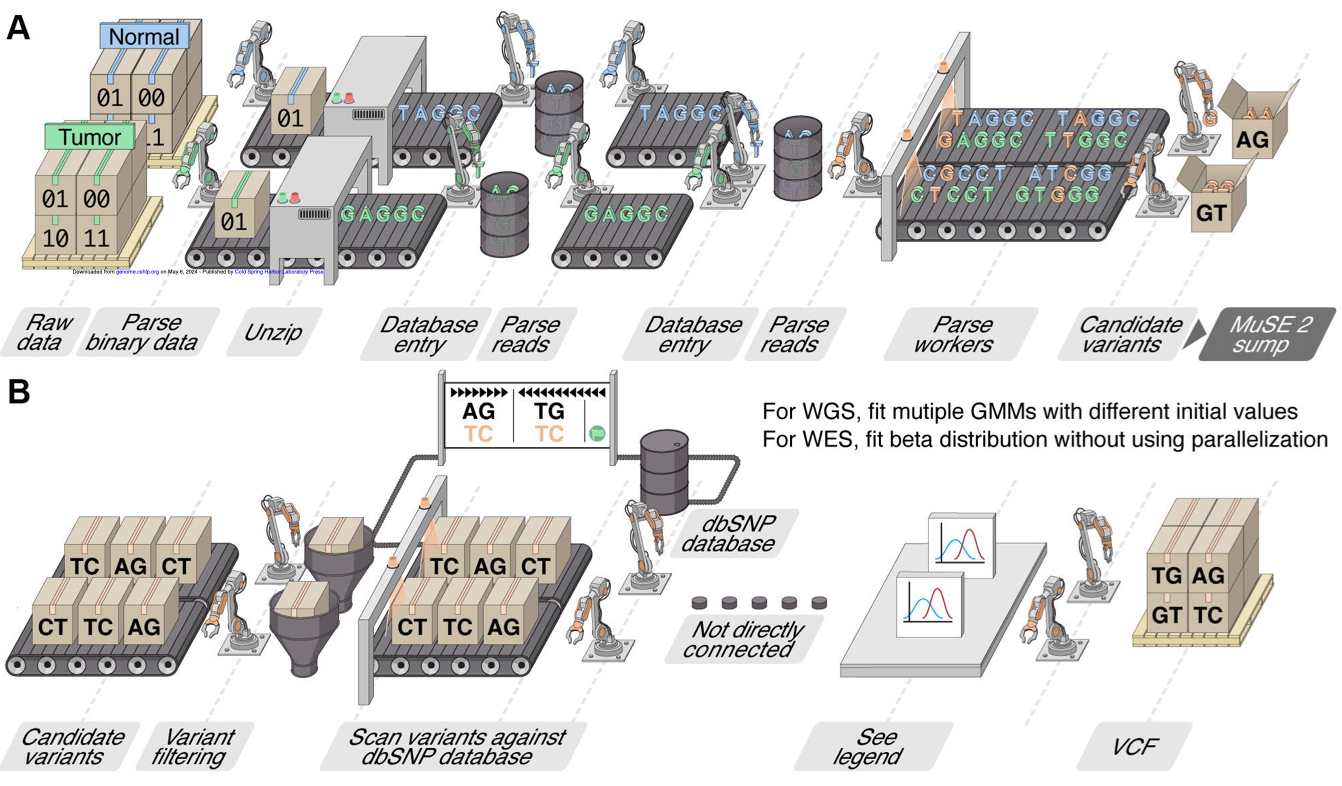
large-scale analysis. My lab is addressing this with [MuSE2], a fast and efficient
mutation
calling method that facilitates large dataset analysis and advances precision
medicine. We
are also interested in improving methods for reconstructing subclonal structures,
which are
critical for understanding cancer evolution and treatment resistance. Our effort on
developing software tools like [CliPP] helps overcome
limitations in previous methods by
significantly reducing computational resources and time, through penalized likelihoods [Characterizing
ITH]. These advancements are critical to understanding intratumor
heterogeneity and cancer evolution, providing important evidence for translational research
to improve patient outcomes.
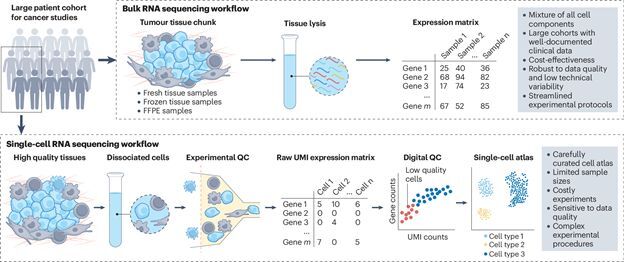
Tissues, including tumors, contain diverse cell types, each with unique transcriptional
patterns that can be studied through RNA expression data. While Single-cell RNA
sequencing (scRNA-seq) provides detailed insights, it is often costly and
challenging for large-scale
use. Bulk RNA-seq is more affordable but mixes signals from different cell types. To
address
this, deconvolution methods like [DeMixSC] help separate
these signals, improving analysis
of cell proportions and disease mechanisms. In cancer research, deconvolution differentiates
tumor from non-tumor cells, offering insights into pathways, prognosis, and heterogeneity
[DeMixT]. We further
developed an
integrative transcriptomic/genomic deconvolution method to
calculate [TmS]
(tumor-specific total mRNA expression), a feature of cancer cell plasticity,
with a striking ability to predict prognosis across cancers. Spatial transcriptomics
data builds on this by adding another dimension, preserving the spatial arrangement of cells
to
help
map tumor microenvironments (TME). This spatial context provides crucial insights into how
cells
interact within their environments, which is essential for understanding tumor progression.
We recently developed DeMixNB to characterize spatial distributions of tumor-specific
gene
expression. By integrating bulk, single-cell, and spatial data, we can achieve deeper
insights, advancing more effective and personalized cancer treatment strategies.
Cancer Risk Modeling (TP53) using machine learning and Bayesian models
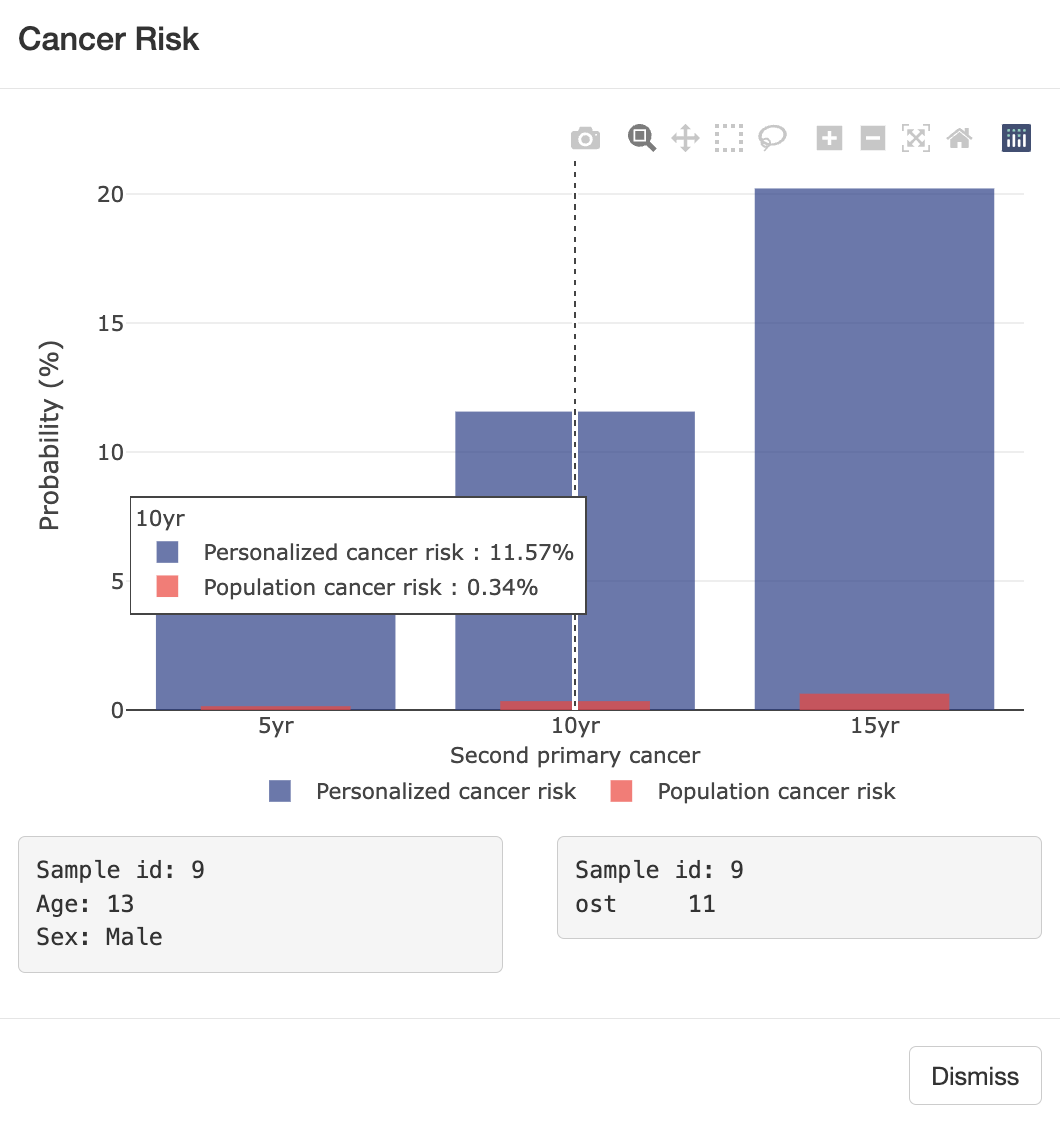
Cancer survivors represent a fast-growing yet under-studied population with respect to cancer
risk, particularly for second primary cancers, which frequently occur in survivors of breast
and bladder cancer. Current risk assessments often overlook prior cancers due to limitations
in large databases like SEER, which mainly account for age and sex. To address this, my lab
studies patients with Li-Fraumeni syndrome (LFS), a hereditary condition
linked
to higher cancer risk. LFS patients often develop multiple primary cancers, offering a
unique opportunity to study cancer risk while accounting for additional factors like
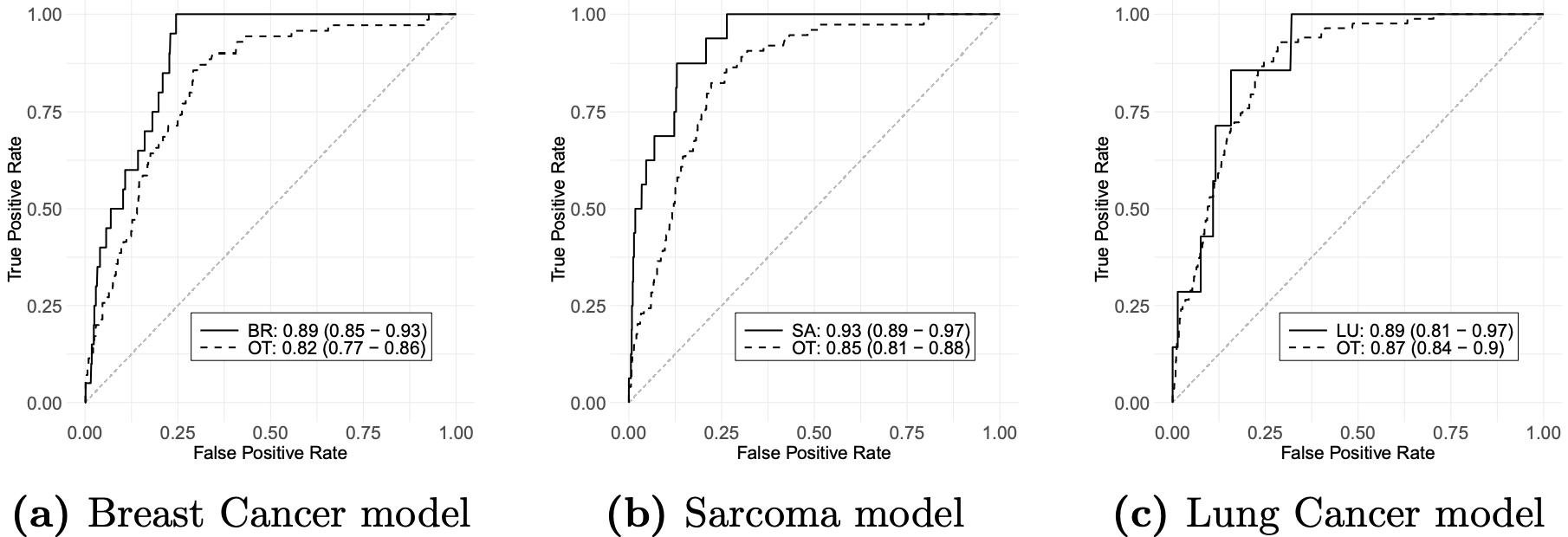
mutation status. Using LFS data, we developed [LFSPRO] to predict both
first and second primary tumors in LFS families. These insights can help physicians and
genetic counselors provide personalized treatment and screening plans, aiming for early
detection of cancers in survivors and LFS patients' Personalized Risk Prediction.
We are also particularly interested in the biological annotation of TP53 mutations,
as the
germline mutations of TP53 are the main cause of LFS. Known as the “guardian of the
genome”,
the TP53 gene plays a critical role in cell signaling, apoptosis, metabolism, DNA
repair and
transcription, and in the meantime it is the most frequently mutated gene in human cancer.
We developed Survival-based clustering of predictors [SCP] using penalized likelihoods
for
survival outcomes, to cluster hundreds of TP53 missense mutations in terms of their
associated early, medium and late onset of cancer in LFS. This research aims to uncover new
patterns in cancer susceptibility and improve predictive models, offering deeper insights
into the genetic underpinnings of cancer risk in LFS patients.
PI: Wenyi Wang
Department of Bioinformatics and Computational Biology
Wenyi Wang (王文漪), Professor, Department of Bioinformatics and Computational Biology, Division of Basic Science Research, The University of Texas MD Anderson, Cancer Center, Houston, Texas
Curriculum Vitae