Latest News

Our immune profiling paper is published in the prestigious JASA!
Our collaborative work, "Immune Profiling Among Colorectal Cancer Subtypes Using Dependent Mixture Models,"
is now published in the Journal of the American Statistical Association.
In this study, we developed a novel Bayesian modeling approach to compare T cell subtypes between early- and late-onset
colorectal cancer. The model identifies immune cell populations that are condition-specific or shared, helping uncover
mechanisms linked to tumor progression and potential treatment strategies.
Congratulations to Yunshan and Shuai!

DeMixSC paper is published at Genome Research!
DeMixSC, an innovative deconvolution approach that overcomes the technological discrepancy between bulk and sc/snRNA-seq data using an improved wNNLS framework. It achieves high accuracy and generalizability, requiring only a small tissue-matched benchmark dataset for the targeted large bulk cohorts.
Free access at https://genome.cshlp.org/content/35/1/147
Exciting Updates from Stats Up AI
We’re thrilled to share two major milestones for Stats Up AI:
1.ASA Approval:
The Stats Up AI interest group has been officially approved by the
American Statistical Association, recognizing its mission to foster
collaboration and innovation at the intersection of statistics and AI.
2.Harvard Data Science Review Publication:
Our recent event, Stats and AI- A Fireside Conversation, was a great
success. A summary of the discussion has been accepted for publication
in the Harvard Data Science Review, further amplifying its impact.
These achievements highlight the growing influence of Stats Up AI in
advancing
the integration of statistics and AI. Stay tuned for future updates as we
continue to engage with this inspiring community!

Congratulations to Carissa for winning the ABRCMS Presentation Award!
Congratulations to our summer intern, Carissa Fong, for winning the presentation award at ABRCMS (Annual Biomedical Research Conference for Minoritized Scientists)! Her award-winning presentation showcased machine learning approaches to effectively predict tumor-specific mRNA expression (TmS). We are so proud of her achievement!

Wang Lab Postdoc Ankita Paul Awarded MD Anderson IDSO Fellowship
Congratulations to our postdoc Ankita, who has been awarded the MD Anderson Institute for Data Science in Oncology (IDSO) Fellowship! This fellowship is a great opportunity that provides junior researchers with advanced training in applying data science to oncology. We are excited to see the impactful contributions she will make through this program!

Wenyi Hooded PhD Graduates Nam and Yunjie(Jeffery) at Rice Univerisity Commencement
Congratuations again to Dr. Nguyen and Dr. Jiang. Wish you all the best and may your future endeavors be filled with success and fulfillment!
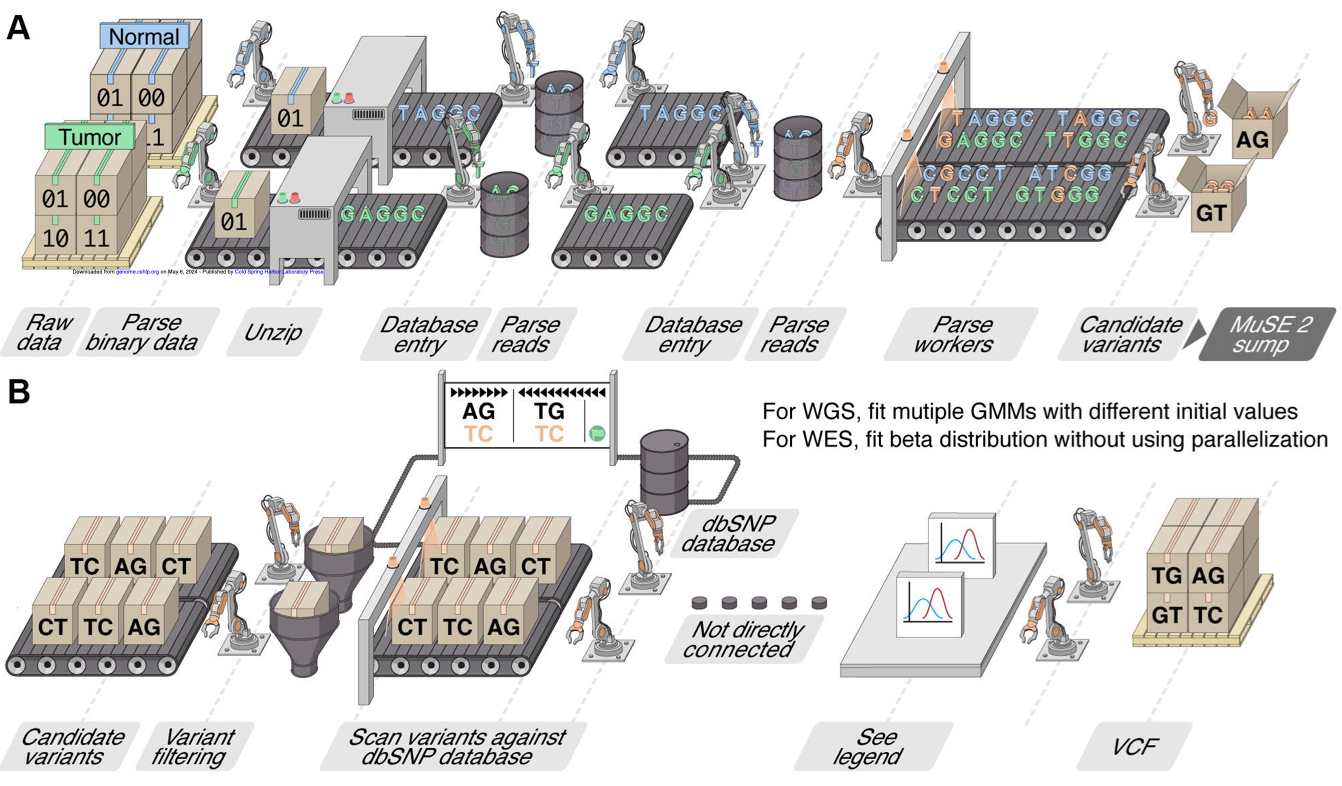
MuSE2.0 paper is online at Genome Research online!
We are exctied to officially introduce MuSE2.0, which reduces computing time by up to 50x compared to MuSE 1.0 and 8-80x compared to other popular callers. Our benchmark study suggests combining MuSE2.0 and the recently accelerated Strelka2 achieves high efficiency and accuracy in analyzing large cancer genomic datasets.
Free access here
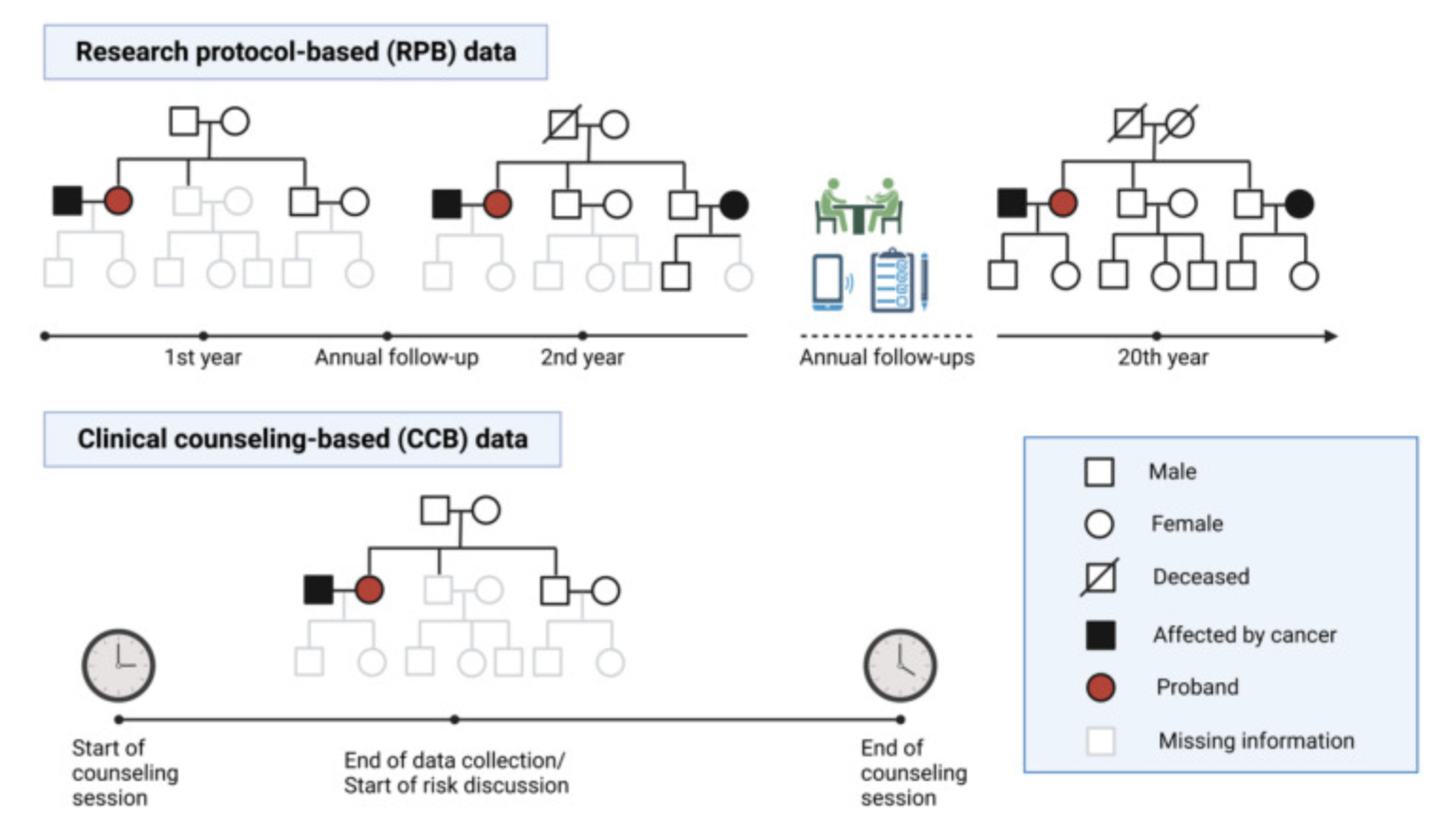
Journal of Clinical Oncology paper is online!
We conducted a validation study of our LFSPRO software suite, which was developed for risk predictions in families with Li-Fraumeni syndrome, on a clinical patient cohort collected as part of the Clinical Cancer Genetics program at MD Anderson Cancer Center. Unlike research datasets that are meticulously collected over decades for research purposes, our unique dataset closely resembles what genetic counselors observe in real counseling sessions. The validation results indicate that our risk prediction models have the potential to assist decision making in clinical settings, and further highlight the importance of such validation in bridging the gap between methodology research labs and clinics.
Free access at https://ascopubs.org/doi/10.1200/JCO.23.01926

Congratulations to Dr. Nguyen and Dr. Jiang!
Congratulations to Nam H Nguyen and Yujie Jiang for successfully
defending
their PhD theses and earning their doctorates!

Don't miss out! Join the Upcoming Webinar: "Empowering Statistics in the Era of AI - A Fireside Conversation"

STATS UP AI - A community for Statistics and Biostatistics
Professors from multiple universities initiate StatsUpAI, aiming
to
elevate
the role of statisticians in AI research.
This movement emphasizes empowering statisticians to lead and
innovate in
addressing real-world challenges through AI research.
As one of the founding members, Wenyi's involvement highlights
her
commitment to advancing statistical methodologies in the era of
artificial
intelligence.
To learn more about StatsUpAI, visit their webpage at
https://statsupai.org
LFSPROShiny paper is published online!
LFSPROShiny is an interactive R/Shiny application designed to
perform risk
prediction and visualization for Li-Fraumeni syndrome (LFS),
a genetic disorder associated with TP53 mutations, enabling
genetic
counselors to assess patient risk profiles and support informed
decision-making without the need for
programming expertise.
Free access at
https://ascopubs.org/doi/10.1200/CCI.23.00167.

Join us on the 2023 Leading Edge of Cancer Research Symposium hosted by MD Anderson on Nov 16-17!
This in-person event provides an incredible no-cost opportunity
to
engage
with and
learn from national and international leaders in cancer
research,
including
an
opportunity to present new research at our poster session.
Ten of the top posters will be chosen for presentations as part
of
the
symposium
agenda as well as monetary awards. Deadline to submit an
abstract is
Oct
20. Please take your lab and program trainees
to
join us!
Click for more details.

Congratulations to the launch of Institute for Data Science in Oncology(IDSO) at MD Anderson!
IDSO integrates the most advanced computational and data science
approaches
with the
institution’s extensive scientific and clinical expertise,
aiming to
profoundly
enhance patient lives by revolutionizing oncological care and
research.
Dr. Wang's lab is proudly affiliated with this pioneering
initiative,
dedicated
to advancements in cancer care and research through innovative
methodologies.
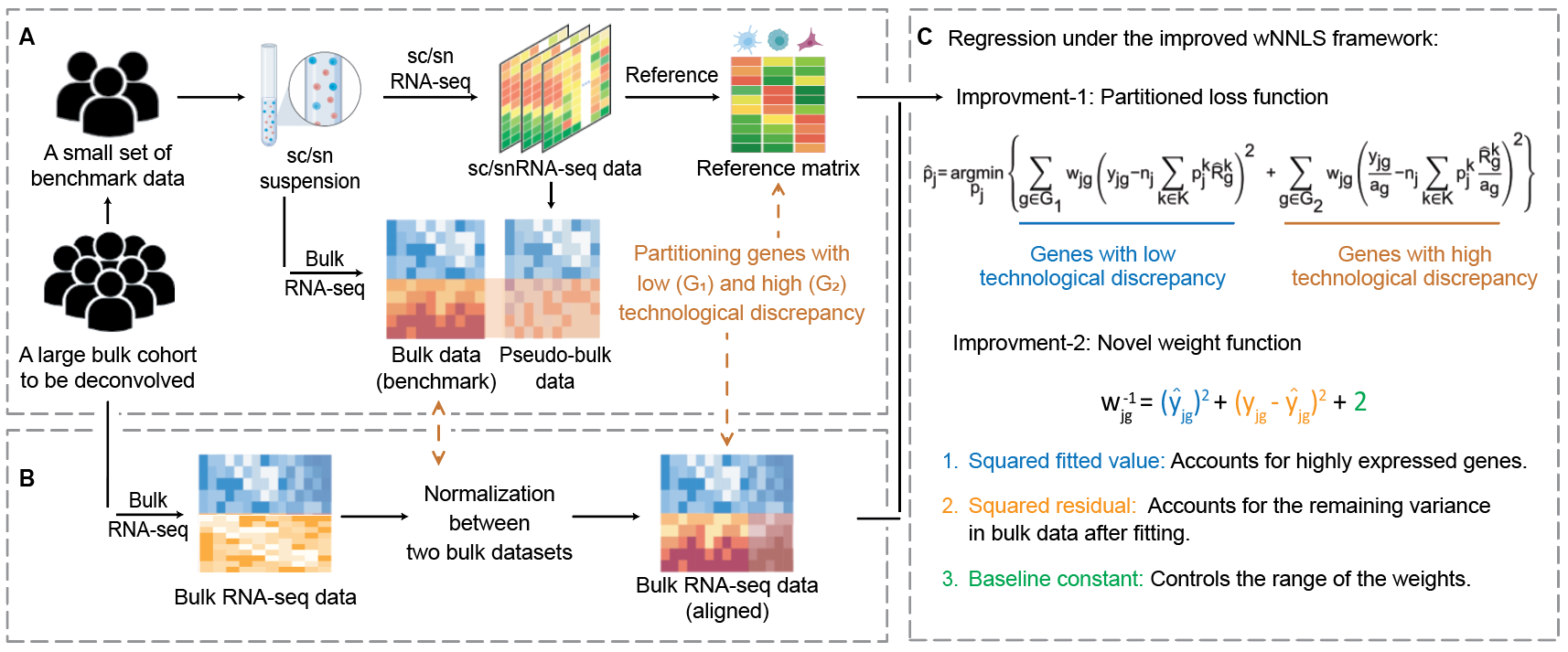
DeMixSC paper is on bioRxiv!
The difference in technology between bulk and sc/snRNA-seq data
significantly
diminishes the accuracy of current deconvolution methods. To
address
this
issue,
we've introduced DeMixSC,
an innovative deconvolution approach that overcomes this
challenge
using an
improved
wNNLS framework. DeMixSC is distinguished by its accuracy in
deconvolution
and its
generalizability to be applied to any large bulk cohorts.
All it requires is a small set of benchmark dataset that match
the
tissue-type of
the targeted large bulk cohorts.
Click for software tool and paper preprint.

Congratulations to our summer interns for their successful Poster Exhibition!
From left to right: Annabel Settle (Intern), Shuai Guo (PhD student), Armina Fani (Intern), Liyang Xie (Intern)
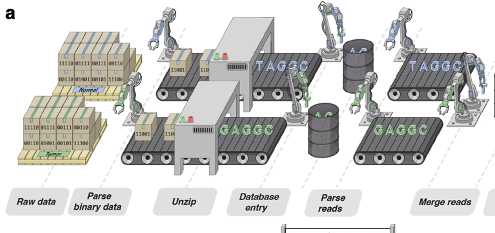
The MuSE2.0 benchmark study is on bioRxiv!
Excited to announce that we have benchmarked MuSE2.0 for somatic mutation
calling in
computing time: finishing one pair of WGS sample < 1 hour, and in
accuracy:
achieves 99% recall of the PCAWG mutations. MuSE 2.0 employs a
multithreaded
producer-consumer model and the OpenMP library for parallel computing.
Our
prepring
is on bioRxiv: https://doi.org/10.1101/2023.07.04.547569. We are looking
for
user
feedbacks.
MuSE2.0 is freely downloadable at
https://github.com/wwylab/MuSE.

Join the 14th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM-BCB) at Houston Sep 3-6 2023!
Wenyi is co-chairing the program committee of the 14th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM BCB 2023), the flagship conference of the ACM SIGBio. It will be held in Houston, TX during September 3-6, 2023, after the past 13 ones held in Atlanta, Boston, Chicago, Newport Beach, Niagara Falls, Online (due to COVID19), Orlando, Seattle, and Washington DC. ACM BCB 2023 will continually showcase leading-edge R&D on data collection, processing, analysis, and knowledge modeling for biological, clinical, and healthcare applications from bench to bedside.
TmS Shiny app online!
In order to facilitate biological correlative analysis using
tumor-specific
tumor-cell total mRNA expression (TmS) across cancer types, we present a
Shiny
app for visual inspections of sequencing data from ~6,500 cancer
patients
without prior programming knowledge. We are looking for user feedbacks:
https://wwylab.github.io/TmS/articles/shinyapp.html.
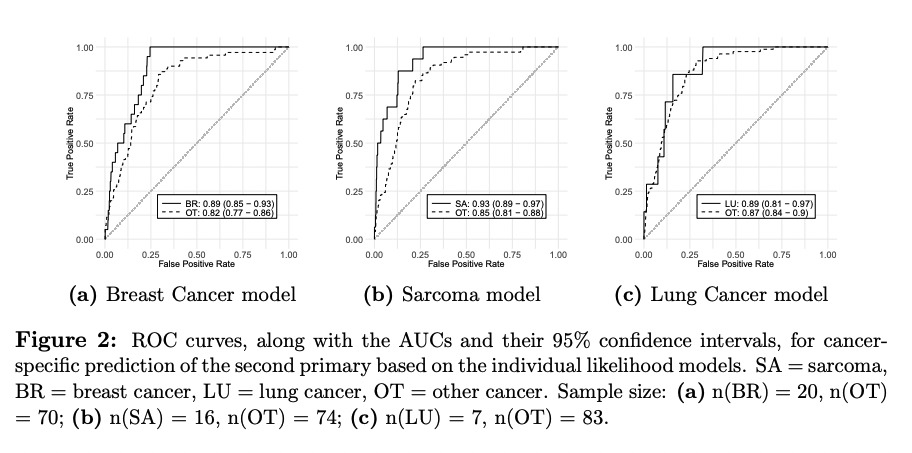
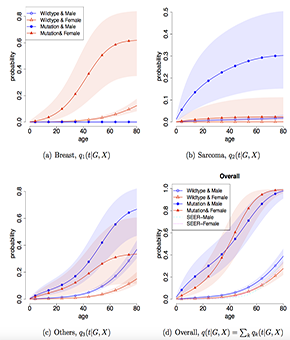
Our new risk prediction modeling paper is on bioRxiv!
There will be more than 20 million cancer survivors in the US by 2026. To help characterize the risk trajectories of this under-studied cancer population, we propose a Bayesian semiparametric model that integrates competing cancer outcomes with a non-homogenous poisson process for recurring events. Check out our preprint for more exciting results we achieved with this model: 10.1101/2023.02.28.530537v2.

Congratulations to collaborator Di Zhao and our dream team for winning the PCF Challenge Award!

Congrats: 4th year PhD student Hoai Nam Nguyen for winning the ASA Section in Lifetime Data Science (LiDS) student paper award!

TmS paper is now in print in Nature Biotechnology November issue.
https://www.nature.com/nbt/volumes/40/issues/11#Features
Exciting TmS News in the media
Check out two recently published complementary reports regarding our
work on
tumor cell total mRNA expression from the Scientist Magazine and
GenomeWeb
Precision Oncology! Click on this title to open both articles together!
Note: If you are only seeing one article, check your
pop-up
blocker
TmS Blog is online
The TmS blog provides a "behind the paper" insight into the motivation
and the
work done. Click the link below to check out the blog.
https://bioengineeringcommunity.nature.com/posts/tumor-specific-total-mrna-expression-a-robust-and-prognostic-feature-across-cancers
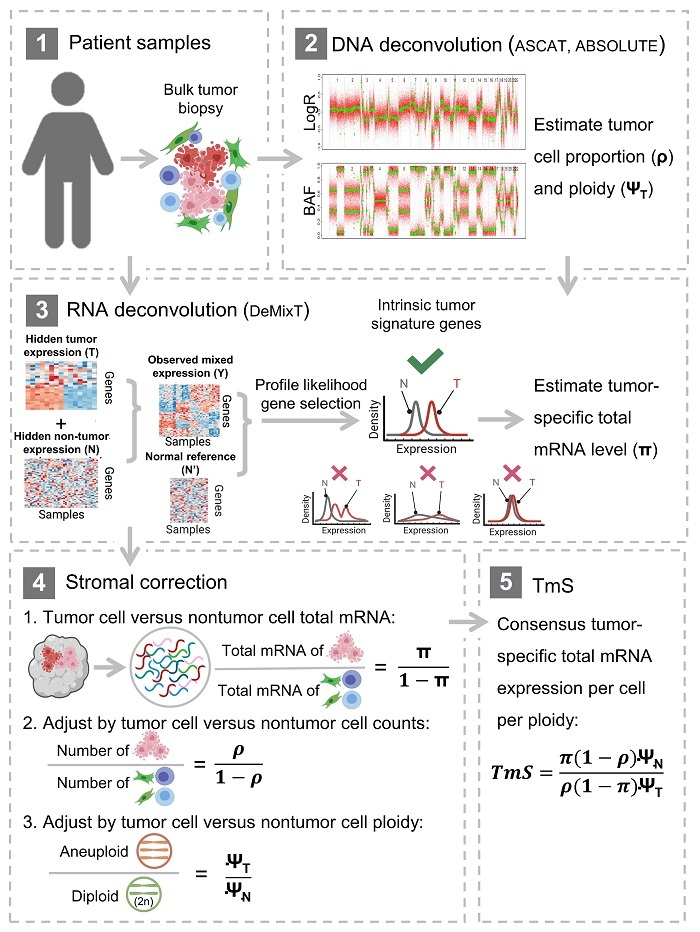
Our TmS Metric is Published at Nature Biotechnology
Very excited to share our paper in Nature Biotechnology today! Huge
amount of
work by Shaolong Cao, Jennifer Wang, Shuangxi Ji et al! It is amazing to
work
with many experts across cancers. Every cancer tells its own story and
our
metric TmS can quantify it! Special thanks to Peter Van Loo's lab for a
wonderful collaboration! Here is the link for the paper. The TmS data
can be
found at https://github.com/wwylab/TmS.
Doi: 10.1038/s41587-022-01342-x

Wenyi will give a keynote talk at RECOMB May 22-25th 2022
The program of RECOMB 2022 is out, click here for more info. This is one of the two major annual conferences in field of computational biology. Wenyi will talk about "Deciphering cancer cell evolution and ecology" at 8am on Wed May 25th!

R01 grant achieved a 3% score!
Our methods R01 grant achieved a 3 percentile priority score from the National Cancer Institute (NCI). This grant titled "statistical methods for analysis of heterogeneous tumors" will support the development of integrative deconvolution models that unite the transcriptomic and genomic aspects of tumor heterogeneity and evolution.

Lab receives DoD Prostate Cancer Research Program Research Program Data Sciencec Award!
Wenyi's lab is awarded a CDMRP Department of Defense Prostate Cancer Research Program Data Science Award to develop an integrated genomic definition and therapeutic strategy for androgen-indifferent prostate cancer, in partnership with Dr. Ana Aparicio from the Department of Genitourinary Medical Oncology at MDACC.

Congratulations to Nam for winning the ASA Statistical Genetics and Genomics Paper Award competition!
The title of the paper is Bayesian estimation of a joint semiparametric recurrent event model of multiple cancer types with application to the Li-Fraumeni Syndrome.
Former PhD student Zeya Wang's paper on Bayesian Edge Regression in Undirected Graphical Models is accepted by JASA
He is now a Machine Learning Engineer at Tik Tok, California. Congrats Zeya!

Wenyi will join the editorial board of JASA Applications and Case Studies as Associate Editor on Jan 1st 2022
A huge congratulations to Wenyi!
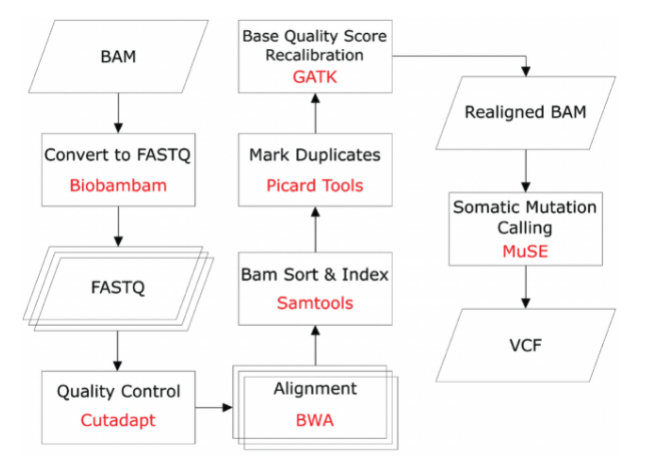
We are proud to announce MuSE2.0
for somatic mutation calling, with 50x speedup from MuSE1.0
Wang lab is on the MD Anderson News!
For our work on genetic diversity within tumors to understand cancer evolution
Congrats: 3rd year PhD student Yujie Jiang received the best paper award from ASA Section in Statistical Genetics and Genomics!
For his work on CliP: fast subclonal architecture reconstruction forcancer cells from genomic DNA sequencing data.
We are excited to introduce a mathematical model to measure an essential RNA feature in tumor cell!
See how we use it to track tumor phenotypes at the link to the preprint below!

Wenyi received a CPRIT grant for cancer prevention as co-PI. Congrats!!
Improving Risk Prediction for Li-Fraumeni Syndrome: A Practical Tool for Clinical Health Care Providers (Banu Arun/Wenyi Wang) - $896,896
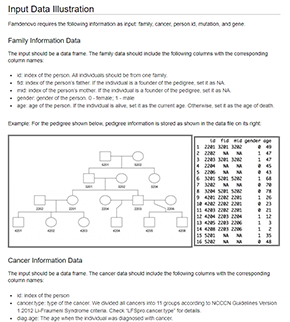
Famdenovo is on Genome Research August Issue
Congratulations to Fan, Elissa, Carlos and Matt!!

A big congrats to Wenyi for promotion to the position of a full professor effective 9/1!
DeMixT 1.2.3 released!
Available on Bioconductor and GitHub
Our two companion papers on LFS are accepted at Cancer Research!
TP53 mutation associated cancer-specific, or multiple-primary-cancer onset penetrances.

Welcome! New postdoctoral fellow Dr. Shuangxi Ji.
Dr. Shuangxi Ji received a PhD in Biological Sciences from University of Birmingham.
Awarded! CZI Seed Network Fund for Human Cell Atlas
We are member of the Retina team.
DeMixT is on Bioconductor.
Recommended download from Bioconductor

MD/PhD student Carlos Vera Recio received NLM fellowship Congrats!
National Library of Medicine (NLM) Training Program in Biomedical Informatics and Data Science
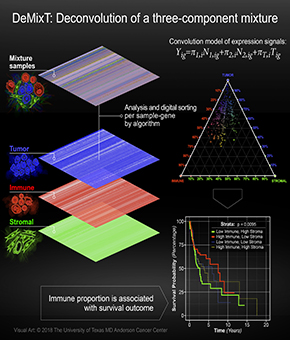
DeMixT paper accepted!
Transcriptome Deconvolution of Heterogeneous Tumor Samples with Immune Infiltration will be published by iScience!
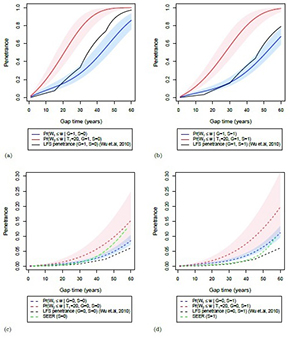
Our multiple primary model accepted by Biostatistics!
Our Bayesian estimation of a semiparametric recurrent event model for estimation of multiple primary cancers paper has been accepted by Biostatistics!
A new subclonal clustering method
Accompanying PCAWG11, our lab has developed a new consensus clustering for subclonal reconstruction software, CSR (pronounced "Caesar").
PCAWG Results
A huge endeavor of the Pan-Cancer Analysis Working Group (PCAWG): Portraits of genetic intra-tumour heterogeneity (ITH) and subclonal selection across cancer types.
LFS statistical model accepted
Our first statistical modeling work for the Li-Fraumeni Syndrome (LFS) is acceptd by Journal of the American Statistical Association after a 3-year journey! Congrats everyone!

Postdocs opportunities
My TAMU collaborator Val Johnson and I are jointly recruiting postdocs in cancer bioinformatics through this 2-year training program at TAMU. Citizenship or green card required. Send us your CV if interested.